16 Past, Present, and Future of West African Sorghum Improvement: Building a Roadmap for Climate-adaptive, Farmer-adopted Varieties
Abstract
Food security and sustainable economic prosperity are key goals for the countries of West Africa (WA). Sorghum (Sorghum bicolor) is among the most important staple crops in WA and has adapted to diverse environments and cultures over many years. However, the process of adaptation is not perfect, and the environment and stakeholder demand is changing, so further sorghum improvement is necessary. National and international sorghum breeding programs are working to improve yield, abiotic stress resilience, and biotic stress resilience. However, it has proved difficult to retain essential end-users’ traits, such as grain quality, and adoption of new varieties has remained low throughout the region. The natural genetic variation within locally adapted and preferred cultivars is key to rapidly develop new climate-adaptive varieties by continuously changing allele frequency and directly selecting outstanding lines through efficient genomics-assisted breeding. It is critical to understand the genetic architecture of adaptive traits, develop genomic tools for marker-assisted and genomic selections, and establish effective data management systems across the region. In this chapter, we describe the genetic diversity of the sorghum germplasm, as well as present an overview of sorghum improvement and perspectives for delivering rapidly and sustainably climate-adaptive breeding products in WA.
Keywords: sorghum, West Africa, breeding, adaptation, genetic diversity
The Sorghum Crop
Sorghum (Sorghum bicolor L. Moench), a C4 grass used mainly for grain and biomass production, is a staple crop for over 500 million people in Africa and Asia (National Research Council, 1996) and grown across various climates (Harlan & de Wet, 1972). Sorghum is a diploid species with 10 pairs of chromosomes and is a predominantly a self-pollinating crop, though it will readily cross-pollinate; gene flow is also commonly seen in sorghum (Barnaud et al., 2008b). The small size of the sorghum genome (about 730 Mbp) makes it a good model system among grass species (McCormick et al., 2018; Paterson et al., 2009) for genomic studies and genomics-assisted breeding.
Cultivated sorghums belong to Sorghum bicolor subspecies. bicolor, with their domestication estimated about 8,000 years ago in East Africa (around present-day Ethiopia and Sudan), followed by their diversification before diffusing south, east, and west (Harlan & de Wet, 1972; Wendorf et al., 1992). A second center of domestication was proposed in West Africa (WA) for guinea margaritiferum (Deu et al., 1994; Folkertsma et al., 2005). Five basic botanical types and ten intermediate types have been defined based on the spikelet and panicle morphology (Harlan & de Wet, 1972). These types—bicolor, durra, guinea, caudatum, and kafir—are originally distributed with respect to geographic regions across Sub-Saharan Africa. These types are used by sorghum breeders worldwide at different levels based on their agronomic importance.
Additionally, four wild races of sorghum have been defined: virgatum, aethiopicum, arundinaceum, and verticilliflorum; these predominate in northeastern Africa, the semi-arid, tropical, and savanna Sahelian regions of West Africa, respectively (de Wet, 1978). These races are grouped into S. bicolor subspecies verticilliflorum (Harlan & de Wet, 1972). In sorghum, wild relatives are rarely used for crop improvement (Ananda et al., 2020), perhaps due to the great diversity available in the domesticated gene pool, compared to crops such as wheat, where wild relatives are often used as donors of traits such as disease resistance (Gill et al., 2011). Still, West African wild, weedy, and admixed sorghums may be useful sources of favorable alleles for stress resilience traits if suitable alleles cannot be identified in the domesticated germplasm (Tesso et al., 2008).
Regardless of botanical type, almost all traditional varieties (landraces) of sorghum in West Africa are tall (2–4 m) and photoperiod-sensitive; that is, they require a critical minimum day-length to flower. Guineas, characterized by an open panicle and resistance to pathogen infection, are predominantly distributed in the humid savannas of West Africa. Caudatums are highly appreciated by breeders, mainly due to their high yield potential (ISRA, 2005), especially in Senegal. Durras, with a compact panicle and usually curved peduncle, are more adapted to the semi-arid Sahelian zone and usually used in breeding programs to contribute drought resilience traits.
Sorghum in West Africa
1. Cropping Systems and Uses
In West Africa, sorghum is mostly cultivated in environments characterized by limited soil fertility, drought, heavy bird attacks, and phosphorus- and aluminum-limited soils (Leiser et al., 2015), where other cereals such as maize and pearl millet can hardly survive. Almost all sorghum production in the region depends on rain-fed farming systems, along with lack of agricultural inputs and vulnerability to climate change, with drought acting as the most common climate factor that limits the production (Mundia et al., 2019). The diversity of farming systems and cultural influences, such as human consumption of sorghum grain for the preparation of meals, including couscous, lakh, To, nieling, mony, and local beer, are key factors for sorghum production (Barnaud et al., 2008a, 2008b). Grain quality for food consumption and farmers’ preferences remain valuable parameters that determine the types of sorghum varieties grown by smallholder farmers. It has been suggested that locally focused studies need to be undertaken to plan for climate-adaptive varieties to meet the needs of the growing population (Mundia et al., 2019). However, the development of broadly adaptive and resilient varieties would contribute to sustainable food security globally.
2. Genetic Diversity
The genetic diversity of West African sorghum is extremely high and is structured by botanical type, with several ancestral populations within botanical type across countries (Deu et al., 2006; Deu et al., 2008; Faye et al., 2019, 2021b; Folkertsma et al., 2005; Maina et al., 2018; Olatoye et al., 2018). However, several gaps exist in the national and international ex situ collections of WA with untapped genetic diversity and less accessibility to the global sorghum community (Upadhyaya et al., 2017). The French colonial science agency ORSTOM (Office de la recherche scientifique et technique outre-mer), predecessor to the current IRD (Institut de Recherche pour le Développement) made collections of sorghum landraces in Senegal, Nigeria, Niger, and Mali in the 1970s (Borgel & Séquier, 1977; Clément & Houdiard, 1977). These collections exist in several genebanks (IRD, ICRISAT, USDA-NPGS, etc.) and constitute sources of adaptive traits for breeding programs. In national breeding programs, most of the duplicated seed lots of these collections have been lost due to poor conservation. More recent collections have been assembled to partly complete the existing gaps and are available to the global sorghum community as ex situ collections. The existing germplasm conservation must remain a key activity to limit the loss of diversity in small and young breeding programs.
Sorghum local varieties harbor genetic variation that should be favorable for developing climate-adaptive varieties (Meyer & Purugganan, 2013). Sorghum is one of the most genetically diverse crops due to its extensive diversification, local adaptation across multiple agroclimatic zones, and continuing wild-crop gene flow that occurred after its domestication (Sagnard et al., 2011). This diversity presents great opportunities, such as the identification of climate-resilient and biotic stress resistant sources (Faye et al., 2021a); however, the diversity of WA sorghum poses challenges such as understanding local adaptation and recovering adaptive traits across various environments. Advances in sequencing and high-throughput genotyping, such as genotyping-by-sequencing (Elshire et al., 2011; Poland et al., 2012) can generate cost-effective and high-density SNP (single nucleotide polymorphism) markers. The development of high-throughput genotyping, combined with large-scale phenotyping, helps to identify natural variants controlling adaptive traits (Faye et al., 2021a; b).
Sorghum Improvement in West Africa
West African smallholder farmers have selected and diffused adapted sorghum for thousands of years through mass selection and seed exchange. Unfortunately, this history of sorghum improvement by smallholders remains poorly understood. Early sorghum breeding in francophone West Africa, prior to independence, was conducted by IRAT (Institut de recherches agronomiques tropicales et des cultures vivrières), a French colonial institute. The sorghum improvement program was based in Bambey, Senegal but was part of a research network across French colonial West Africa for germplasm and multi-environment trials. After independence, the national breeding programs of francophone countries of West Africa, (Institut Sénégalais de Recherches Agricoles–ISRA in Senegal, Institut National de la Recherche Agronomique du Niger–INRAN in Niger, Institut d’Economie Rurale–IER in Mali, Institut Togolais de Recherche Agronomique–ITRA in Togo, and Institut de l’Environnement et de Recherches Agricoles–INERA in Burkina Faso) were established to continue the work of IRAT. These programs work in close collaboration with international breeding programs to develop or test varieties for diverse environments. In later sections, we will focus on the history of the formalized sorghum improvement programs in Senegal and Niger.
1. History of Sorghum Improvement in Senegal
Sorghum breeding in Senegal reflects sorghum breeding in WA and has yielded several good varieties that were developed for specific agro-ecological regions. Table 1 depicts the main sorghum varieties that were released and disseminated to farmers for the different agro-ecological zones of Senegal. However, considerable extension efforts must be made to disseminate these varieties in all smallholder farmers and cropping systems.
| Varieties | Code | Year of Release | Maturity Cycle (days) | Seed Type | Grain Yield (ton ha-1) |
Agro-ecological Zone
|
| Payenne | ISRA-S-618-1 | 2015 | 85–90 | White creamy Semi-vitreous | 2.5–3.5 |
North peanut basin (300–600 mm)
|
| Golobe | ISRA-S-618-2 | 2015 | 85–90 | White creamy Semi-vitreous | 2.5–3.5 |
North peanut basin (300–600 mm)
|
| CE180-33 | CE180-33 | 1983 | 90 | White floury | 2–3 |
Centre-north (300–600 mm)
|
| CE151-262 | CE151-262 | 1980 | 90 | White creamy Semi-vitreous | 4–5 |
Senegal river valley
|
| CE196-7-2-1 | CE196-7-2-1 | 1983 | 95–100 | White creamy Semi-vitreous | 3–3.5 |
Centre-west (Mbour area)
|
| CE145-66 | CE145-66 | 1980 | 100 | White floury | 2.5–3 |
Centre-north, Sine-Saloum, South-east
|
| Darou | ISRA-S-622B | 2011 | 105 | White creamy Semi-vitreous | 2.5–3 |
South peanut basin (600–800 mm)
|
| Faourou | ISRA-S-621B | 2011 | 105 | White creamy Semi-vitreous | 2.5–3 |
South peanut basin (600–800 mm)
|
| F2-20 | F2-20 | 1983 | 110 | White creamy Semi-vitreous | 3–3.5 |
Centre-south, South, and South-east
|
| Nguinthe | ISRA-S-621A | 2011 | 110 | White creamy Semi-vitreous | 2–3 |
South-east and North Casamance (800–1000mm)
|
| Nganda | ISRA-S-622A | 2011 | 110 | White creamy Semi-vitreous | 2 – 3 |
South-east and North Casamance (800–1000mm)
|
1.1 Senegalese Sorghum Cropping Systems
Sorghum is cultivated in Senegal in three different cropping systems. The most important, in terms of production, is rainfed cultivation (sowing in July) in the southern groundnut basin, eastern Senegal, and middle and upper Casamance. The two other production systems are flood recession cultivation (décrue) from November to April along the Senegal River valley (Roy, 2008) and irrigated cultivation during the rainy season (sowing in July) and cold off-season (sowing in October) in the Senegal River valley. Some varietal improvement efforts were carried out in each of the three different cropping systems. Three basic botanical types (guinea, caudatum, and durra) and their intermediate types are predominantly cultivated.
1.2 Beginning of Formal Sorghum Improvement Activities
There have been several phases of sorghum improvement in Senegal (Figure 1). Sorghum genetic improvement started in the 1950s, but it was not until 1960 that defined breeding targets were established for sorghum varietal improvement (Mauboussin et al., 1977). Before 1960, the objective was to focus on farmers’ preferences by selecting sorghum lines phenotypically, similar to local cultivars, with better productivity and grain quality. Between 1960 and 1965, sorghum lines were selected more intensively. Breeding objectives were still focused on general grain yield improvement while obtaining varieties close to local cultivars. Short stature was not systematically sought, and trials were carried out without fertilizer inputs. A mass selection activity in the local cultivars of the flood recession and rainfall cropping systems was carried out on the Senegal River. This period also marked the beginning of the development of sorghum hybrids in Senegal. However, the hybrid program was not continued consistently, and no sorghum hybrids were released in Senegal (ISRA, 2005; FAO, 2012).
Figure 1
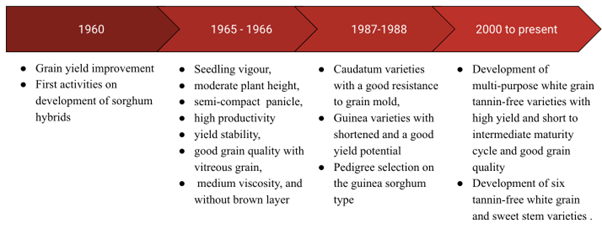
1.3 1960s–1980s: Landrace Purification and Green Revolution-style Varieties
Starting in 1965–1966, several initiatives were undertaken, including the improvement of farming practices, the introduction of founder cultivars from the world collection, the addition of a section for diversification in the Senegal River area, and, above all, the definition of precise selection targets and objectives by the agro-ecological region (Mauboussin et al., 1977). Generally, common selection objectives were defined, then specific objectives were added for each area considered. These common objectives, which broadly correspond to “Green Revolution” style varieties (Swaminathan, 2006), included (a) moderate plant height (~1.8–2 m for rainfed system, ~1.5 m for irrigated system); (b) seedling vigor; (c) semi-compact and erect panicle with good exsertion; (d) high productivity and yield stability; (e) good grain quality with vitreous endosperm, medium viscosity of grain flour, and without brown layer, tannin, and anthocyanin. Specific objectives were also defined for different cropping zones: lines with short maturity cycles less than 90 days, good dryness, and resistance to grain mold for the central zone of the peanut basin; lines with intermediate maturity cycle between 100–110 days and resistance to grain mold for the south-central zone of the peanut basin; lines with late maturity cycle between 110–120 days and resistance to grain mold for the south-eastern Senegal and northern Casamance; and hybrids and lines with early maturity cycle between 80–90 days, very productive, and short stature of about 1.80m for the Senegal River zone.
Selection activities between 1970 to 1980 focused on development of early maturity lines and hybrids for the rainfed and irrigated systems (ISRA, 2005; Luce, 1988). Some of the key varieties that were developed during this period are presented in Table 1. Among them, IRAT 204 (also known as CE 151-262) was developed in 1980 from a cross between IRAT 11 (CE 90) and 73-71 (IS 12610), an Ethiopian line. This variety was developed particularly for the Soudano-Sahelian agro-climate zone (annual rainfall of 500–700 mm) and has been used in several breeding programs in WA because of its high tolerance to biotic and abiotic stresses, such as drought, leaf disease, and insect damage (Muleta et al., 2021).
1.4 1980s–1990s: Refocusing Breeding Targets for Caudatum and Guinea Pure lines
In 1987–1988, the breeding target was reoriented because of the difficulties of the dissemination and adoption of the improved caudatum varieties relative to traditional varieties belonging mainly to the guinea type (ISRA, 2005; Luce, 1995). A study conducted in 1987 and 1988 showed that local guinea cultivars were generally less susceptible to grain mold than improved caudatum varieties, had good adaptive traits, good resistance to diseases and insects, and good grain quality, but their productivity was generally low and their height high. It was therefore proposed to conduct two breeding targets for the Center-South and South zones, with the objectives to (a) emphasizing obtaining caudatum varieties with a good resistance to grain mold and (b) obtaining guinea varieties with short stature and good yield potential while maintaining the characteristics of traditional cultivars (FAO, 2012; ISRA, 2005; Luce, 1988). The year 1988 marked the beginning of pedigree selection on the guinea sorghum type. A recurrent selection program was also initiated for the creation of populations between guinea and caudatum types. In 1993, the selection program for the irrigated system in the Senegal river was stopped.
1.5 2000s–2010s: Focus on Multi-purpose Caudatum Varieties for Central Senegal
From 2000 to 2018, the program focused on the development of multi-purpose, white grain, tannin-free varieties with high yield potential, short to intermediate maturity cycle, and good grain quality that could be used for both food and feed. A cross between the varieties IRAT 204 and Sorvato-1, a line with good grain quality from Togo, resulted in the development and release of six pureline caudatum type varieties (Nguinthe, Faourou, Nganda, Darou, Payenne, and Golobe), all tannin-free, white grain, and sweet stems (FAO, 2012). These varieties were successfully used in bread making to produce compound bread (20% sorghum flour and 80% wheat flour). Their vegetative part, highly prized by animals, is stored after harvest and used as fodder during the dry season. The average adoption rate of the improved varieties in Senegal has increased from low (5%) for the varieties released in the 1980s (Trouche et al., 1999) to moderate (25%) for varieties released in the 2010s (Ndiaga Cisse, personal communication, March 2018). This current adoption rate is comparable to the 27.4% average adoption rate of improved sorghum cultivars in Sub-Saharan Africa (Walker & Alwang, 2015). Two of the major factors limiting the adoption of improved varieties is the lack of financial resources and material to apply the full technology package, as well as the fact that improved varieties do not meet farmers’ grain quality preferences. The hybrid program initiated in the 1960s was also resumed in 2012. Recently, ten hybrids were developed, five for the south peanut basin zone and five for the north zone, all of which are in the registration process (Diatta et al., 2019, 2021).
1.6 2020s–Onward: Focus on Breeding Adaptive Varieties for Particular Agro-climates
Since 2018, emphasis has been put on defining and using breeding product profiles (BPPs) based on end-users’ preferences and adaptation criteria to guide breeding program objectives.
In the Senegal River, farmers preferred traits consist of short stature, more precocity (<90 d) than the elite improved varieties, and the presence of awn on the panicle to prevent bird attacks (Bullard, 1988; Perumal & Subramaniam, 1973). The current varieties, Golobe and Payenne, are medium-stature and early-maturing. The first breeding target is to improve the presence of awn on the panicle. The second target is to develop varieties that are insensitive to the cool-off season because high stature varieties tend to reduce their height during the cool season.
In the central and south agro-ecological zones, the main challenges are related to crop management practices; thus, the goal is to deliver varieties that are adapted to the length of the growing season (e.g., moderate photosensitive varieties), or early (< 90 d), medium (95-105 d), and late (> 110 d) maturing varieties for a given zone. Photoperiod sensitivity is an adaptive trait where cycle durations of sensitive varieties coincide with growing season lengths, allowing for local adaptation to abiotic (water limitation) and biotic (grain mold and bird attacks) constraints (Curtis, 1968; Kouressy et al., 2008). In addition, the open panicle trait and grain hardness are targeted as proxies to reduce grain mold damage in caudatum varieties in the central and southern regions. The variety Grinkan from the Mali breeding program is used to bring semi-open panicle, seedling vigor, and moderate photoperiod sensitive traits into elite varieties. In parallel, more efforts are being put into guinea sorghum types for the sorghum growing regions with annual rainfall >800 mm by combining elite caudatum varieties and local guineas to develop varieties with vitreous and hard grains.
To increase the adoption rate of sorghum varieties, current breeding activities involve gathering farmers’ preferences, starting with end-users’ demands, and integrating farmers in the process of varietal selection (Aissata, 2018; Sissoko et al., 2019). These demands are formulated as BPPs for the different agro-ecological zones. To develop new versions of farmers’ preferred cultivars, backcross populations are being developed to introgress the lgs-1 striga resistance allele from the donor line SRN39 (Gobena et al., 2017), staygreen alleles from the donor line B35, grain mold resistance, and high grain protein digestibility into elite varieties using marker-assisted backcross (MABC).
2. History of the Nigerien Sorghum Breeding Program
2.1 Role of Niger within the French Colonial System
Sorghum breeding activities started earlier with the French institute IRAT in the 1960s across WA through screening and evaluations of local varieties in Niger. Promising varieties were selected for yield improvement and varietal purification for broad adaptation using either mass selection or pure line selection (Chantereau et al., 2002). The French institute, Office de la Recherche Scientifique et technique Outre-mer (ORSTOM), which become the Institute of Research for Development (IRD) collected also sorghum landraces, across Niger in 1976 and 2003 with national partners (Bezançon et al., 2009). Local varieties such as Jan Jare, Bogoba, Babadia Fara, and Babadia Ja were used as screening material from Niger. The objectives were to develop high yielding varieties with medium height and adapted to semi-arid regions. The variety IRAT 204 (early maturing at 90 days) was among the promising varieties developed for WA, which represents 2.74% of the total sorghum cultivated area in Niger. Moreover, IRAT 11 (known as L30, derived from line 137-62 and Jan Jare) was selected for medium maturity (110 days) in the 1970s. However, L30 was not adopted by farmers due to its low food quality.
2.2 Launching of the INRAN National Program
INRAN was created in 1975 following IRAT programs in Niger with four regional research centers in the country based at Kollo, Maradi, Niamey, and Tahoua. The objectives were to contribute to food security and rural development in Niger under the ministry of agriculture. Sorghum breeding is among the earlier programs of other crops in Niger attempting to develop high yielding varieties tolerant to stresses. Major targeted traits were early maturity, acceptable food quality, and good nutritional value. Caudatum and durra-caudatum were the types adapted for semi-arid regions.
2.3 1980s–2010s: INRAN Collaborations with INTSORMIL and ICRISAT
Sorghum breeding in Niger started with germplasm exchange through international programs. Early generations and advanced generations from existing US parents were introduced. Several breeding nurseries were conducted and germplasm exchanged during the International Sorghum and Millet Collaborative Research Support Program (INTSORMIL) to establish strong and durable selection programs in Niger. Sorghum hybrid production started with germplasm from international collaborations for joint evaluations with breeding programs from Purdue University and the International Crops Research Institut for the Semi-Arid Tropics (ICRISAT). Progenies developed by ICRISAT were evaluated in Niger through the Sorghum Elite Progeny Observation Nursery (SEPON). From 1991 to 1995, two sorghum varieties were released: the variety SEPON 82 (M 90038) and SRN39 (ICSV 1007 BF) (Bantilan et al., 2004). Moreover, germplasm exchanges with international partnerships lead to the development of hybrid varieties, such as the NAD1 hybrid (ATx623 x MR732), which was released for the Sahelian agro-climatic zone in 1991 (Ministère de l’Agriculture du Niger, 2012). The NAD1 hybrid is a white seeded and medium-maturity line that yields over two tons per hectare, higher than the local check Mota Maradi (Kapran et al., 1997, 2007). However, the developed varieties were either not broadly adapted or less appreciated by end-users especially during food preparation. NAD1, SRN39, and SEPON 82 represent 0.45%, 0.07%, and 4.95% respectively of the total sorghum cultivated area in Niger in 2009 (Walker & Alwang, 2015).
2.4 2020s–Onward
Nowadays, the Niger breeding program aims to focus more on understanding the needs of end-users through Participatory Rural Appraisal (PRA) and market needs, while accelerating breeding cycles to deliver new varieties with better appreciation (Aissata, 2018). The current breeding targets are defined as BPPs to release adapted lines for different regions across the country. The main breeding targets involve breeding for high yielding varieties with tolerance to drought stress; resistance to striga, downy mildew, midge, and long smut; early maturing; and high grain and forage quality.
Genetic and Genomic Resources for West African Sorghum Breeding
The sorghum germplasm in WA, including landraces, were (and are still being) adapted to various environmental variations and farming practices for many generations; thus, selection must have favored the fixation of adaptive loci (Orr, 1998) useful for breeding (Leiser et al., 2014). The majority of elite varieties in breeding programs of WA were developed over 30 years ago. Though some of these varieties might be obsolete, several of them are still adapted to different environments across WA. For example, IRAT 204, SEPON 82, Sorvato-1, Sariaso 11, Sariaso 15, Grinkan, Mota Maradi, and SRN39 are key elite breeding lines. These key varieties have been extensively used to generate crosses across breeding programs, and it is expected that the presence of shared common haplotypes across diverse breeding populations may facilitate deciphering the genetic control of key traits and their pyramiding in improved varieties.
1. Genome-wide Genotyping of Genebank Collections
The thousands of West African sorghum accessions available in genebanks are potentially useful sources of trait donors for pre-breeding (Kimber et al., 2013; Upadhyaya et al., 2017). Recently, using sequencing technologies, several genebank collections have been characterized, providing useful resources for breeding programs of the region (Bellis et al., 2020; Billot et al., 2013; Faye et al., 2019; Lasky et al., 2015; Maina et al., 2018; Olatoye et al., 2018). In WA, national and international sorghum collections were genotyped using high-density SNP markers that are publicly available to the global scientific community (Faye et al., 2019, 2021b; Maina et al., 2018; Olatoye et al., 2018). Genomic analyses showed several ancestral populations structured hierarchically with respect to botanical types and agro-climatic zones across countries.
A West African sorghum association panel (WASAP) was assembled by breeders, physiologists, and geneticists in Senegal, Niger, Togo, and Mali and genotyped using GBS (Faye et al., 2021b). Based on 159,101 high-quality biallelic SNPs obtained for this panel, genetic characterization showed that high genetic diversity exists in sorghum across WA where 8 ancestral populations are found in Senegal, Niger, Mali, and Togo germplasms. The genetic characterization of the WASAP and different collections, ex-situ, allowed to better understand the genetic diversity, population structure, putative alleles for local adaptation, and farmer selections existing in WA. These genomic resources for WA sorghum germplasm would contribute to the genetic dissection of adaptive traits and development of new varieties adapted to the current and future environmental conditions.
2. The Potential of Genome-wide Association Studies for Trait Discovery
Cultivars harboring favorable alleles for local adaptation are potential donors of winning traits and can be used for pre-breeding through genomics-assisted breeding. Genome-wide association studies (GWAS) help to identify natural variations that control key traits in adapted cultivars and multi-parental populations. GWAS have been successfully applied in sorghum panels including WA cultivars, with the identification of known and novel loci associated with photoperiodic flowering, plant height, drought tolerance, grain pigmentation, farmer preferences, and climatic variables (Bellis et al., 2020; Faye et al., 2019, 2021b; Hu et al., 2019; Lasky et al., 2015; Maina et al., 2018; Olatoye et al., 2018). The Maturity genes, Ma6, SbCN8, and SbCN12, have been mapped several times using different sorghum panels of WA, suggesting the predominant role of these genes for photoperiodic flowering differences in the WA sorghums. However, the strong population structure observed in the germplasm suggests applying regional GWAS on specific populations or on multiparental populations with balanced gene pools.
The backcross-nested association mapping (BC-NAM) populations developed by the Mali breeding program (Teme, 2018), Australia breeding program (Jordan et al., 2011), and the sorghum MAGIC population from Purdue University (Ongom & Ejeta, 2017) were shown to segregate for key adaptive agronomic traits, including high yield, drought tolerance, early maturity, grain quality, high protein digestibility, and disease and pest resistance. These multiparental populations constitute useful genetic resources for GWAS and varietal improvement in WA (Teme, 2018). Also, following a similar strategy, nested-association mapping populations (NAM) were developed in Senegal and Niger to constitute useful genetic resources for breeding programs.
Perspectives on New Breeding Technologies for Sorghum in West Africa
1. Overview of Adoption of New Technology: Separating Fads from Solutions
Technology in plant breeding and genetics is constantly evolving. Crop improvement programs must carefully consider which new technologies they adopt or not (Bernardo, 2016b ; Simmonds, 1991). Some technologies may be useful for some programs and not for others, or for specific breeding targets within a program and not others. All breeding programs, from well-financed commercial breeding to small developing country NARS programs, must consider the breeding technology to use depending on the status of their breeding program and the genetic architecture of priority traits. For instance, a simulation of a Haitian sorghum breeding showed that genomic selection may, or may not, be more cost-effective than phenotypic selection depending on the underlying genetic architecture for yield variation (Muleta et al., 2019). This highlights the need to integrate effective selection methods for small NARS programs. Genomics-assisted breeding based on shared genetic and genomic resources can help to leverage limitations of field phenotyping for optimal selection in early generations across multiple environments (Cobb et al. 2018; Deshpande et al., 2016).
The development of genomic tools contributes to solving the roadblocks that limit or delay the process of achieving breeding product concepts using conventional breeding. Conventional breeding has yielded many elite varieties of important economic values. However, phenotypic selection in early generations for many adaptive traits, such as photoperiodic flowering and drought tolerance, is difficult for small and young breeding programs. One of the reasons is that multiple field trials are generally required for optimum selection. Crop variety update is a slow process in small and young breeding programs and can take up to 10 years from first crossing to releasing new varieties to farmers and end-users. This process can take even longer when considering large-scale farmer diffusion and adoption, while a limited attention is given to most staple food crops such as sorghum in Africa. With advances in next-generation sequencing technologies and cost-effective genotyping platforms, there are opportunities to modernize small and young breeding programs to accelerate varietal development and continuously release new founder lines through genomics-assisted breeding (Poland, 2015). Figure 2 resumes the key local and elite germplasm useful for the development of climate-adaptive varieties for smallholder farmers and end-users using genomics-assisted breeding.
Figure 2
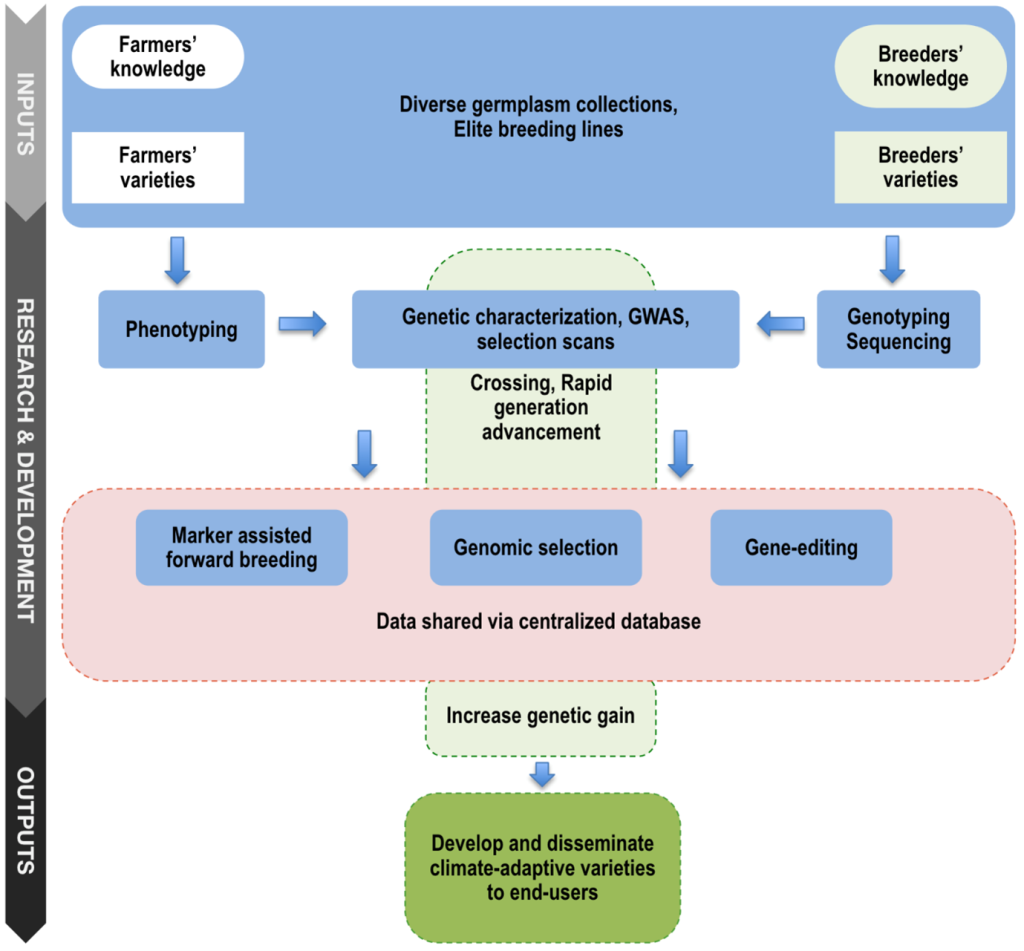
2. Rapid Generation Advancement and Targeted Use of SNPs
Rapid generation advancement has been proposed to accelerate variety product profile development in the International Rice Research Institute (IRRI) breeding program (Collard et al., 2019). The method consists of rapidly fixing genotypes to homozygosity with 3.5 to 4 generations per year and starting yield trials with F5-derived lines for seed increase instead of using F7 lines. In this context, quality and uniformity control of seed lots and crosses can also contribute to establishing an efficient breeding product development. In the case of sorghum in WA, a defined set of several SNP markers can be used for quality control of any pair or multiple founder breeding lines. Having highly polymorphic markers will complement the available trait-associated cost-effective Kompetitive Allele Specific PCR (KASP) markers to establish an efficient marker-assisted selection (MAS) program for forward breeding of simply inherited traits. Quality control activities of WA breeding programs can be carried out at the regional facility established at ISRA/CERAAS. However, out-sourcing genotyping in specialized laboratories would be more effective for large-scale genotyping, especially for saving breeders’ time and focusing on main breeding activities.
3. Systems for Data Management and Sharing
Enabling data sharing and developing a centralized database that stores phenotypes, genotypes, and pedigree breeding information will contribute to establishing an efficient genomics-assisted breeding pipeline in WA. Since 2014, the Breeding Management System (BMS) of the Integrated Breeding Platform (IBP) has actively helped the management of breeding activities of several crops, including sorghum, in several NARS in WA. Such a centralized database would be particularly beneficial for GS model training to improve prediction accuracy and have access to correlated phenotypic and genotypic data from different environments. These datasets would contain high frequency of common haplotypes shared across populations and a wide range of environmental trials. The more related data and larger training populations available to train GS models, the better the prediction accuracy as previously demonstrated in the International Maize and Wheat Improvement Center (CIMMYT) wheat breeding (Battenfield et al., 2016). Accessible breeding and training populations will be potentially useful to develop climate resilient varieties because of the extensive agro-climate variation and local environment testing that were exercised by breeders in the region through the IAVAO network.
4. Genomic Selection
Genomic selection approach was first established in animal breeding and is an efficient method to accelerate breeding cycling and increase genetic gain (Jannink et al., 2010). Its usage in crop breeding programs is recent and has shown promising results, especially in crops such as maize, wheat, and barley (Michel et al., 2017; Pierre et al., 2016; Zhang et al., 2017). Simulations and empirical studies of GS have demonstrated a potential to increase genetic gain while reducing the number of breeding cycles in several food crops (Battenfield et al., 2016; Bernardo, 2016a). In GS, the whole performance of individuals is assessed regardless of interaction between positive and negative alleles by estimating the individual total performance based on individual genomic-estimated breeding values (GEBV), thus more efficient when dealing with polygenic traits. Both genomic selection and MAS provide advantages to more rapidly increasing genetic gain per unit time and unit cycle, and therefore can be integrated together in the breeding pipeline for more efficient breeding.
Applications of GS are currently limited in small breeding programs, including WA sorghum breeding. Most NARS breeding programs run elite by exotic breeding rather than elite by elite breeding compared to most advanced public or private international breeding programs. However, correlated genotypes exist in diverse West African sorghum populations because the genetic diversity is structured by botanical types and subpopulations within botanical type across countries (Faye et al., 2021b) and diverse key founder lines such as IRAT204, Sorvato-1, Grinkan, SEPON 82, and Sariaso 11 are shared among breeding programs. Continuous sharing of breeding materials across breeding programs will be useful for effective GS implementation.
5. Gene Editing
CRISPR/Cas9, Cas12a, and Cas13 gene editing systems are emerging technologies that hold opportunities to increase genetic gain in breeding. Integrating CRISPR-based gene editing with genomic-assisted selection may help to purge deleterious alleles while favoring rapid genetic gain. This system yields potential interest to deal with negative epistasis traits (Sackman & Rokyta, 2018). Important traits such as tannin content, which may be negatively associated with grain mold resistance (GMR) and grain quality, or grain yield with plant height could involve negative epistasis. The condensed tannin in the pigmented testa of sorghum seed is controlled by the presence of loss-of-function alleles at both Tannin1 (Tan1) and Tannin2 (Tan2) genes (Wu et al., 2012, 2019), and indirectly involved in GMR (Ackerman et al., 2021; Nida et al., 2021), which can be directly edited in tannin-free elite varieties. Other loci such as YELLO SEED1 (Y1) and Y3 genes have also shown to contribute to GMR (Nida et al., 2019) and thus can be edited to develop resistant varieties.
Conclusion
Important progress has been achieved in assembling and genetically characterizing the WA sorghum germplasm. High genetic diversity exists within and across breeding programs, with multiple ancestral populations shared across countries. To take advantage of the diverse sorghum germplasm, interdisciplinary researchers must work in synergy to understand sorghum adaptation to specific and across diverse environments. Genomics-assisted breeding through population genomics, GWAS in multiparental populations, MAS, GS, and gene-editing can offer the ability to directly trace the untapped diversity and introduce natural variation into locally preferred cultivars. However, a rigorous scientific approach, such as the GoHy method, must be employed to avoid wasting time, resources, or distracting breeding activities as a result of adopting new technologies without proper setting of development goals and rigorous hypotheses. The rigorous definition of BPPs and GoHy will not only allow testing hypotheses for functional causal variants for adaptive traits but also increase genetic gain to establish food security in West Africa. As phenotyping and genotyping platforms continue to be more and more sophisticated, it is urgent to enable continuous data sharing and management with a centralized database.
Acknowledgements
Portions of this manuscript have been adapted from the Kansas State University PhD dissertations of J.M.F. and F.M. This study is made possible by the support of the American People provided to the Feed the Future Innovation Lab for Collaborative Research on Sorghum and Millet through the United States Agency for International Development (USAID) under Cooperative Agreement No. AID-OAA-A-13-00047. The contents are the sole responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government.
References
Ackerman, A., Wenndt, A., & Boyles, R. (2021). The sorghum grain mold disease complex: Pathogens, host responses, and the bioactive metabolites at play. Frontiers in Plant Science, 12. https://doi.org/10.3389/fpls.2021.660171
Aissata, M. I. (2018). Characterization of sorghum production constraints and ideal plant and variety traits as perceived by farmers in Niger. JSM Biotechnology & Biomedical Engineering, 5(1), 1084.
Ananda, G. K. S., Myrans, H., Norton, S. L., Gleadow, R., Furtado, A., & Henry, R. J. (2020). Wild sorghum as a promising resource for crop improvement. Frontiers in Plant Science, 11. https://doi.org/10.3389/fpls.2020.01108
Bantilan, M. C. S., Deb, U. K., Gowda, C. L. L., Reddy, B. V. S., Obilana, A. B., & Evenson, R. E. (2004). Sorghum genetic enhancement: Research process, dissemination and impacts. International Crops Research Institute for the Semi-Arid Tropics.
Barnaud, A., Joly, H. I., McKey, D., Deu, M., Khasah, C., Monné, S., & Garine, E. (2008a). Gestion des ressources génétiques du sorgho (Sorghum bicolor) chez les Duupa (Nord Cameroun). Cahiers Agricultures, 17(1), 178–182. https://doi.org/10.1684/agr.2008.0170
Barnaud, A., Trigueros, G., McKey, D., & Joly, H. I. (2008b). High outcrossing rates in fields with mixed sorghum landraces: how are landraces maintained? Heredity, 101, 445–452. https://doi.org/10.1038/hdy.2008.77
Battenfield, S. D., Guzmán, C., Gaynor, R. C., Singh, R. P., Peña, R. J., Dreisigacker, S., Fritz, A. K., & Poland, J. A. (2016). Genomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding program. The Plant Genome, 9(2). https://doi.org/10.3835/plantgenome2016.01.0005
Bellis, E. S., Kelly, E. A., Lorts, C. M., Gao, H., DeLeo, V. L., Rouhan, G., Budden, A., Bhaskara, G. B., Hu, Z., Muscarella, R., Timko, M. P., Nebie, B., Runo, S. M., Chilcoat, N. D., Juenger, T. E., Morris, G. P., dePamphilis, C. W., & Lasky, J. R. (2020). Genomics of sorghum local adaptation to a parasitic plant. Proceedings of the National Academy of Sciences, 117(8), 4243–4251. https://doi.org/10.1073/pnas.1908707117
Bernardo, R. (2016a). Genomewide predictions for backcrossing a quantitative trait from an exotic to an adapted line. Crop Science, 56(3), 1067–1075. https://doi.org/10.2135/cropsci2015.09.0586
Bernardo, R. (2016b). Bandwagons I, too, have known. Theoretical and Applied Genetics, 129, 2323–2332. https://doi.org/10.1007/s00122-016-2772-5
Bezançon, G., Pham, J.-L., Deu, M., Vigouroux, Y., Sagnard, F., Mariac, C., Kapran, I., Mamadou, A., Gérard, B., Ndjeunga, J., & Chantereau, J. (2009). Changes in the diversity and geographic distribution of cultivated millet (Pennisetum glaucum (L.) R. Br.) and sorghum (Sorghum bicolor (L.) Moench) varieties in Niger between 1976 and 2003. Genetic Resources and Crop Evolution, 56, 223–236. https://doi.org/10.1007/s10722-008-9357-3
Billot, C., Ramu, P., Bouchet, S., Chantereau, J., Deu, M., Gardes, L., Noyer, J.-L., Rami, J.-F., Rivallan, R., Li, Y., Lu, P., Wang, T., Folkertsma, R. T., Arnaud, E., Upadhyaya, H. D., Glaszmann, J.-C., & Hash, C. T. (2013). Massive sorghum collection genotyped with SSR markers to enhance use of global genetic resources. PLOS ONE, 8. https://doi.org/10.1371/journal.pone.0059714
Borgel, A., & Séquier, J. (1977). Prospection de mils pénicillaires et sorghos en Afrique de l’Ouest: Campagne 1976: Niger. ORSTOM.
Bullard, R. W. (1988). Characteristics of bird-resistance in agricultural crops. Proceedings of the Thirteenth Vertebrate Pest Conference, 62, 305–309. https://digitalcommons.unl.edu/vpcthirteen/62
Chantereau, J., Nabos, J., Poisson, C., & Rouanet, G. (2002). Le mil, le sorgho, le blé et l’orge. L’Institut de recherches agronomiques tropicales et des cultures vivrières 1960-1984, Vol. 1. CIRAD.
Clément, J.-C., & Houdiard, P. (1977). Prospection des mils pénicillaires et sorghos en Afrique de l’Ouest: Campagne 1976: Nigeria-Sénégal. ORSTOM.
Cobb, J. N., Biswas, P. S., & Platten, J. D. (2018). Back to the future: revisiting MAS as a tool for modern plant breeding. Theoretical and Applied Genetics, 132, 647–667. https://doi.org/10.1007/s00122-018-3266-4
Collard, B. C. Y., Gregorio, G. B., Thomson, M. J., Islam, M. R., Vergara, G. V., Laborte, A. G., Nissila, E., Kretzschmar, T., & Cobb, J. N. (2019). Transforming rice breeding: Re-designing the irrigated breeding pipeline at the International Rice Research Institute (IRRI). Crop Breeding, Genetics and Genomics, 1. https://doi.org/10.20900/cbgg20190008
Curtis, D. L. (1968). The Relation Between the Date of Heading of Nigerian Sorghums and the Duration of the Growing Season. The Journal of Applied Ecology, 5(1), 215–226. https://doi.org/10.2307/2401286
Deshpande, S., Rakshit, S., Manasa, K. G., Pandey, S., & Gupta, R. (2016). Genomic approaches for abiotic stress tolerance in sorghum. In S. Rakshit & Y.-H. Wang (Eds.), The Sorghum Genome (pp. 169–187). Springer International Publishing. https://doi.org/10.1007/978-3-319-47789-3_9
Deu, M., Gonzalez-de-Leon, D., Glaszmann, J.-C., Degremont, I., Chantereau, J., Lanaud, C., & Hamon, P. (1994). RFLP diversity in cultivated sorghum in relation to racial differentiation. Theoretical and Applied Genetics, 88, 838–844. https://doi.org/10.1007/BF01253994
Deu, M., Rattunde, F., & Chantereau, J. (2006). A global view of genetic diversity in cultivated sorghums using a core collection. Genome, 49(2), 168–180. https://doi.org/10.1139/g05-092
Deu, M., Sagnard, F., Chantereau, J., Calatayud, C., Hérault, D., Mariac, C., Pham, J.-L., Vigouroux, Y., Kapran, I., Traore, P. S., Mamadou, A., Gerard, B., Ndjeunga, J., & Bezançon, G. (2008). Niger-wide assessment of in situ sorghum genetic diversity with microsatellite markers. Theoretical and Applied Genetics, 116, 903–913. https://doi.org/10.1007/s00122-008-0721-7
Deu, M., Sagnard, F., Chantereau, J., Calatayud, C., Vigouroux, Y., Pham, J.-L., Mariac, C., Kapran, I., Mamadou, A., Gérard, B., Ndjeunga, J., & Bezançon, G. (2010). Spatio-temporal dynamics of genetic diversity in Sorghum bicolor in Niger. Theoretical and Applied Genetics, 120, 1301–1313. https://doi.org/10.1007/s00122-009-1257-1
de Wet, J. M. J. (1978). Systematics and evolution of sorghum sect. Sorghum (Gramineae). American Journal of Botany, 65(4), 477–484. https://doi.org/10.2307/2442706
Diatta, C., Tovignan, T. K., Adoukonou-Sagbadja, H., Aidara, O., Diao, Y., Sarr, M. P., Ifie, B. E., Offei, S. K., Danquah, E. Y., & Cisse, N. (2019). Development of sorghum hybrids for stable yield and resistance to grain mold for the center and south-east of Senegal. Crop Protection, 119, 197–207. https://doi.org/10.1016/j.cropro.2019.02.001
Diatta, C., Sarr, M. P., Tovignan, T. K., Aidara, O., Dzidzienyo, D. K., Diatta-Holgate, E., Faye, J. M., Danquah, E. Y., Offei, S. K., & Cisse, N. (2021). Multienvironment evaluation of tannin-free photoperiod-insensitive sorghum (Sorghum bicolor (L) Moench) for yield and resistance to grain mold in Senegal. International Journal of Agronomy, 2021. https://doi.org/10.1155/2021/5534314
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., & Mitchell, S. E. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE, 6. https://doi.org/10.1371/journal.pone.0019379
FAO. (2012). Catalogue officiel des variétés.
Faye, J. M., Akata, E. A., Sine, B., Diatta, C., Cisse, N., Foncéka, D., & Morris, G. P. (2021a). Quantitative and population genomics suggest a broad role of stay-green loci in the drought adaptation of sorghum. The Plant Genome, 15(1). https://doi.org/10.1002/tpg2.20176
Faye, J. M., Maina, F., Akata, E. A., Sine, B., Diatta, C., Mamadou, A., Marla, S., Bouchet, S., Teme, N., Rami, J.-F., Foncéka, D., Cisse, N., & Morris, G.P. (2021b). A genomics resource for genetics, physiology, and breeding of West African sorghum. The Plant Genome, 14(2). https://doi.org/10.1002/tpg2.20075
Faye, J. M., Maina, F., Hu, Z., Foncéka, D., Cisse, N., & Morris, G. P. (2019). Genomic signatures of adaptation to Sahelian and Soudanian climates in sorghum landraces of Senegal. Ecology and Evolution, 9(10), 6038–6051. https://doi.org/10.1002/ece3.5187
Folkertsma, R. T., Rattunde, H. F. W., Chandra, S., Raju, G. S., & Hash, C. T. (2005). The pattern of genetic diversity of Guinea-race Sorghum bicolor (L.) Moench landraces as revealed with SSR markers. Theoretical and Applied Genetics, 111, 399–409. https://doi.org/10.1007/s00122-005-1949-0
Gill, B. S., Friebe, B. R., & White, F. F. (2011). Alien introgressions represent a rich source of genes for crop improvement. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7657–7658. https://doi.org/10.1073/pnas.1104845108
Gobena, D., Shimels, M., Rich, P. J., Ruyter-Spira, C., Bouwmeester, H., Kanuganti, S., Mengiste, T., & Ejeta, G. (2017). Mutation in sorghum low germination stimulant 1 alters strigolactones and causes striga resistance. Proceedings of the National Academy of Sciences, 114(17), 4471–4476. https://doi.org/10.1073/pnas.1618965114
Harlan, J. R., & de Wet, J. J. M. (1972). A simplified classification of cultivated sorghum. Crop Science, 12(2), 172–176. https://doi.org/10.2135/cropsci1972.0011183X001200020005x
Hu, Z., Olatoye, M. O., Marla, S., & Morris, G. P. (2019). An integrated genotyping-by-sequencing polymorphism map for over 10,000 sorghum genotypes. The Plant Genome, 12(1). https://doi.org/10.3835/plantgenome2018.06.0044
ISRA. (2005). Bilan de la recherche agricole et agroalimentaire au Sénégal. Institut sénégalais de recherches agricoles, 524.
Jannink, J.-L., Lorenz, A. J., & Iwata, H. (2010). Genomic selection in plant breeding: from theory to practice. Briefings in Functional Genomics, 9(2), 166–177. https://doi.org/10.1093/bfgp/elq001
Jordan, D. R., Mace, E. S., Cruickshank, A. W., Hunt, C. H., & Henzell, R. G. (2011). Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Science, 51(4), 1444–1457. https://doi.org/10.2135/cropsci2010.06.0326
Kapran, I., Amadou, M., Abdou, M., Souley, S., Conde, N., Axtell, J. D., Tyler, T., & Ejeta, G. (1997). Heterosis and prospects for marketing sorghum hybrids in Niger. The Genetics and Exploitation of Heterosis in Crops.
Kapran, I., Grenier, C., & Ejeta, G. (2007). Introgression of genes for striga resistance into African landraces of sorghum. Integrating New Technologies for Striga Control (pp. 129–141). World Scientific. https://doi.org/10.1142/9789812771506_0010
Kimber, C. T., Dahlberg, J. A., & Kresovich, S. (2013). The gene pool of sorghum bicolor and its improvement. In A.H. Paterson (Ed.), Genomics of the Saccharinae (pp. 23–41). Springer New York.
Kouressy, M., Traoré, S., Vaksmann, M., Grum, M., Maikano, I., Soumaré, M., Traoré, P. S., Bazile, D., Dingkuhn, M., & Sidibé, A. (2008). Adaptation des sorghos du Mali à la variabilité climatique. Cahiers Agricultures, 17(2), 95-100. https://doi.org/10.1684/agr.2008.0189
Lasky, J. R., Upadhyaya, H. D., Ramu, P., Deshpande, S., Hash, C. T., Bonnette, J., Juenger, T. E., Hyma, K., Acharya, C., Mitchell, S. E., Buckler, E. S., Brenton, Z., Kresovich, S., & Morris, G. P. (2015). Genome-environment associations in sorghum landraces predict adaptive traits. Science Advances, 1(6). https://doi.org/10.1126/sciadv.1400218
Leiser, W. L., Rattunde, H. F. W., Weltzien, E., Cisse, N., Abdou, M., Diallo, A., Tourè, A. O., Magalhaes, J. V., & Haussmann, B.I. (2014). Two in one sweep: aluminum tolerance and grain yield in P-limited soils are associated to the same genomic region in West African sorghum. BMC Plant Biology, 14(206). https://doi.org/10.1186/s12870-014-0206-6
Leiser, W. L., Rattunde, H. F. W., Piepho, H.-P., Weltzien, E., Diallo, A., Toure, A., & Haussmann, B. I. G. (2015). Phosphorous efficiency and tolerance traits for selection of sorghum for performance in phosphorous-limited environments. Crop Science, 55(3), 1152–1162. https://doi.org/10.2135/cropsci2014.05.0392
Luce, C. (1988). L’amélioration variétale du sorgho au Sénégal: Acquis (1950-1986) et perspectives de recherches. Institut Senegalais de Recherches Agricoles, 1(2), 1–22. https://rivieresdusud.uasz.sn/xmlui/handle/123456789/701
Luce, C. (1995). Le Sorgho. Création variétale et étude de la diversité génétique: Synthèse des travaux effectués par le CIRAD-CA 1961-1994. https://agritrop.cirad.fr/325955
Maina, F., Bouchet, S., Marla, S. R., Hu, Z., Wang, J., Mamadou, A., Abdou, M., Saïdou, A.-A., & Morris, G. P. (2018). Population genomics of sorghum (Sorghum bicolor) across diverse agroclimatic zones of Niger. Genome, 61, 223–232. https://doi.org/10.1139/gen-2017-0131
Mauboussin, J.-C., Gueye, J., & N’Diaye, M. (1977). L’amélioration du Sorgho au Sénégal. Agronomie Tropicale XXXll, 32(3), 303–310. https://agritrop.cirad.fr/421362
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., Kennedy, M., Amirebrahimi, M., Weers, B. D., McKinley, B., Mattison, A., Morishige, D. T., Grimwood, J., Schmutz, J., & Mullet, J. E. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. The Plant Journal, 93(2), 338–354. https://doi.org/10.1111/tpj.13781
Meyer, R. S., & Purugganan, M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nature Reviews Genetics, 14, 840–852. https://doi.org/10.1038/nrg3605
Michel, S., Ametz, C., Gungor, H., Akgöl, B., Epure, D., Grausgruber, H., Löschenberger, F., & Buerstmayr, H. (2017). Genomic assisted selection for enhancing line breeding: merging genomic and phenotypic selection in winter wheat breeding programs with preliminary yield trials. Theoretical and Applied Genetics. 130, 363–376. https://doi.org/10.1007/s00122-016-2818-8
Ministère de l’Agriculture du Niger. (2012). Catalogue National des Espèces et Variétés Végétales (CNEV).
Muleta, K., Felderhoff, T., Winans, N., Walstead, R., Charles, J., Armstrong, J., Mamidi, S., Plott, C., Vogel, J., Lemaux, P., Mockler, T., Grimwood, J., Schmutz, J., Pressoir, G., & Morris, G. (2021). The recent evolutionary rescue of a staple crop depended on over half a century of global germplasm exchange. Science Advances, 8(6). https://doi.org/10.1126/sciadv.abj4633
Muleta, K. T., Pressoir, G., & Morris, G. P. (2019). Optimizing genomic selection for a sorghum breeding program in Haiti: A simulation study. G3: Genes, Genomes, Genetics, 9(2), 391–401. https://doi.org/10.1534/g3.118.200932
Mundia, C. W., Secchi, S., Akamani, K., & Wang, G. (2019). A regional comparison of factors affecting global sorghum production: The case of North America, Asia and Africa’s sahel. Sustainability, 11(7), 2135. https://doi.org/10.3390/su11072135
National Research Council. (1996). Lost crops of Africa: Volume I: Grains. National Academies Press.
Nida, H., Girma, G., Mekonen, M., Lee, S., Seyoum, A., Dessalegn, K., Tadesse, T., Ayana, G., Senbetay, T., Tesso, T., Ejeta, G., & Mengiste, T. (2019). Identification of sorghum grain mold resistance loci through genome wide association mapping. Journal of Cereal Science, 85, 295–304. https://doi.org/10.1016/j.jcs.2018.12.016
Nida, H., Girma, G., Mekonen, M., Tirfessa, A., Seyoum, A., Bejiga, T., Birhanu, C., Dessalegn, K., Senbetay, T., Ayana, G., Tesso, T., Ejeta, G., & Mengiste, T. (2021). Genome-wide association analysis reveals seed protein loci as determinants of variations in grain mold resistance in sorghum. Theoretical and Applied Genetics, 134, 1167–1184. https://doi.org/10.1007/s00122-020-03762-2
Olatoye, M. O., Hu, Z., Maina, F., & Morris, G. P. (2018). Genomic signatures of adaptation to a precipitation gradient in Nigerian sorghum. G3: Genes, Genomes, Genetics, 8(10), 3269–3281. https://doi.org/10.1534/g3.118.200551
Ongom, P. O., & Ejeta, G. (2017). Mating design and genetic structure of a multi-parent advanced generation inter-cross (MAGIC) population of sorghum (Sorghum bicolor (L.) Moench). G3: Genes, Genomes, Genetics, 8(1), 331–341. https://doi.org/10.1534/g3.117.300248
Orr, H. A. (1998). The population genetics of adaptation: The distribution of factors fixed during adaptive evolution. Evolution; International Journal of Organic Evolution, 52(4), 935–949. https://doi.org/10.1111/j.1558-5646.1998.tb01823.x
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., Haberer, G., Hellsten, U., Mitros, T., Poliakov, A., Schmutz, J., Spannagl, M., Tang, H., Wang, X., Wicker, T., Bharti, A. K., Chapman, J., Feltus, F. A., Gowik, U., … & Rokhsar, D. S. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature, 457, 551–556. https://doi.org/10.1038/nature07723
Perumal, R. S., & Subramaniam, T. R. (1973). Studies on panicle characters associated with bird resistance in Sorghum. Madras Agricultural Journal, 60, 256–258.
Pierre, C. S., Burgueño, J., Crossa, J., Dávila, G. F., López, P. F., Moya, E. S., Moreno, J. I., Muela, V. M. H., Villa, V. M. Z., Vikram, P., Mathews, K., Sansaloni, C., Sehgal, D., Jarquin, D., Wenzl, P., & Singh, S. (2016). Genomic prediction models for grain yield of spring bread wheat in diverse agro-ecological zones. Scientific Reports, 6(27312). https://doi.org/10.1038/srep27312
Poland, J. (2015). Breeding-assisted genomics. Current Opinion in Plant Biology, 24, 119–124. https://doi.org/10.1016/j.pbi.2015.02.009
Poland, J. A., Brown, P. J., Sorrells, M. E., & Jannink, J.-L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLOS ONE, 7. https://doi.org/10.1371/journal.pone.0032253
Rami, J.-F., Dufour, P., Trouche, G., Fliedel, G., Mestres, C., Davrieux, F., Blanchard, P., & Hamon, P. (1998). Quantitative trait loci for grain quality, productivity, morphological and agronomical traits in sorghum (Sorghum bicolor L. Moench). Theoretical and Applied Genetics, 97, 605–616. https://doi.org/10.1007/s001220050936
Roy, L. (2008). Le sorgho de décrue dans la vallée du Sénégal. https://hal.ird.fr/ird-00179486
Sackman, A. M., & Rokyta, D. R. (2018). Additive phenotypes underlie epistasis of fitness effects. Genetics, 208(1), 339–348. https://doi.org/10.1534/genetics.117.300451
Sagnard, F., Deu, M., Dembélé, D., Leblois, R., Touré, L., Diakité, M., Calatayud, C., Vaksmann, M., Bouchet, S., Mallé, Y., Togola, S., & Traoré, P. C. S. (2011). Genetic diversity, structure, gene flow and evolutionary relationships within the Sorghum bicolor wild–weedy–crop complex in a western African region. Theoretical and Applied Genetics, 123(1231). https://doi.org/10.1007/s00122-011-1662-0
Simmonds, N. W. (1991). Bandwagons I Have Known. Retrieved March 5, 2021, from http://www.sidalc.net/cgi-bin/wxis.exe/?IsisScript=CIMMYT.xis&method=post&formato=2&cantidad=1&expresion=mfn=020016
Sissoko, M., Smale, M., Castiaux, A., & Theriault, V. (2019). Adoption of new sorghum varieties in Mali through a participatory approach. Sustainability, 11(17), 4780. https://doi.org/10.3390/su11174780
Swaminathan, M. S. (2006). An evergreen revolution. Crop Science, 46(5), 2293–2303. https://doi.org/10.2135/cropsci2006.9999
Teme N. (2018, April 8-12). The Malian experience using MARS/ BCNAM for breeding improved sorghum cultivars. Sorghum Conference 21st Century, Cape Town, South Africa.
Tesso, T., Kapran, I., Grenier, C., Snow, A., Sweeney, P., Pedersen, J., Marx, D., Bothma, G., & Ejeta, G. (2008). The potential for crop-to-wild gene flow in sorghum in Ethiopia and Niger: A geographic survey. Crop Science, 48(4), 1425–1431. https://doi.org/10.2135/cropsci2007.08.0441
Trouche, G., Fliedel, G., Chantereau, J., & Barro, C. (1999). Productivité et qualité des grains de sorgho pour le tô en Afrique de l’Ouest: Les nouvelles voies d’amélioration. Agriculture et Développement, 23, 94–107. https://agritrop.cirad.fr/476192
Upadhyaya, H. D., Reddy, K. N., Vetriventhan, M., Ahmed, M. I., Krishna, G. M., Reddy, M. T., & Singh, S. K. (2017). Sorghum germplasm from west and central Africa maintained in the ICRISAT genebank: Status, gaps, and diversity. The Crop Journal, 5(6), 518–532. https://doi.org/10.1016/j.cj.2017.07.002
Walker, T.S., & Alwang, J. eds. (2015). Crop improvement, adoption, and impact of improved varieties in food crops in sub–saharan Africa. CABI. https://doi.org/10.1079/9781780644011.0000
Wendorf, F., Close, A. E., Schild, R., Wasylikowa, K., Housley, R. A., Harlan, J. R., & Królik, H. (1992). Saharan exploitation of plants 8,000 years BP. Nature, 359, 721–724. https://doi.org/10.1038/359721a0
Wu, Y., Guo, T., Mu, Q., Wang, J., Li, X., Wu, Y., Tian, B., Wang, M. L., Bai, G., Perumal, R., Trick, H. N., Bean, S. R., Dweikat, I. M., Tuinstra, M. R., Morris, G., Tesso, T. T., Yu, J., & Li, X. (2019). Allelochemicals targeted to balance competing selections in African agroecosystems. Nature Plants, 5, 1229–1236. https://doi.org/10.1038/s41477-019-0563-0
Wu, Y., Li, X., Xiang, W., Zhu, C., Lin, Z., Wu, Y., Li, J., Pandravada, S., Ridder, D. D., Bai, G., Wang, M. L., Trick, H. N., Bean, S. R., Tuinstra, M. R., Tesso, T. T., & Yu, J. (2012). Presence of tannins in sorghum grains is conditioned by different natural alleles of Tannin1. Proceedings of the National Academy of Sciences of the United States of America, 109(26), 10281–10286. https://doi.org/10.1073/pnas.1201700109
Zhang, X., Pérez-Rodríguez, P., Burgueño, J., Olsen, M., Buckler, E., Atlin, G., Prasanna, B. M., Vargas, M., Vicente, F. S., & Crossa, J. (2017). Rapid cycling genomic selection in a multiparental tropical maize population. G3: Genes, Genomes, Genetics, 7(1), 2315–2326. https://doi.org/10.1534/g3.117.043141

