19 Breeding for Drought Adaptation and Fresh Seed Dormancy of Groundnut in Senegal: Advances, Challenges, and Prospects
Issa Faye, ISRA/CNRA, Senegal
Aïssatou Sambou, ISRA/CERAAS, Senegal
Gualbert Seraphin Dorego, ISRA/CNRA, Senegal
Daniel Foncéka, CIRAD & ISRA/CERAAS, Senegal
Hodo-Abalo Tossim, ISRA/CERAAS, Senegal
Amadou Ka, ISRA/CNRA, Senegal
Thierno Sarr, Gaston Berger University, Senegal
Ousmane Sy, ISRA/CNRA, Senegal
Abstract
Groundnut (Arachis hypogaea L.) is a multi-purpose legume widely cultivated in the semiarid tropics. In Senegal, groundnut is historically an important crop, and it remains one of the commodities with a huge potential for exports. However, due to the country’s location in the Sahelian zone in West Africa, crop production is subject to low and erratic rainfall patterns and unpredictable end of the rainy season. Therefore, groundnut breeding programs were initiated early in the 20th century to develop and release drought-tolerant cultivars with fresh seed dormancy (to avoid rapid sprouting). This review highlights the significant advances made in these research areas to reduce yield losses and groundnut quality deterioration. Challenges concerning the effects of climate change on agricultural production, which will keep increasing over the coming decades, are also addressed herein. Finally, we provided some insights on integrating the new breeding tools and methods such as high-throughput phenotyping, marker-assisted and genomic selection to breed groundnut varieties that are more adapted to the agro-ecologies in Senegal.
Keywords: groundnut, drought, stress, seed dormancy, cultivars, Senegal
Background
In Senegal, the agricultural sector accounts for more than 9.8% of the gross domestic product (ANSD, 2022), with groundnut being the most important cash crop. Groundnut crop also plays a central role in the development of the rural economy. It employs nearly 87% of the working population, positioning Senegal among the 10 largest world producers. However, groundnut productivity in Senegal (1.46 kg ha-1) is still less than the world average yield, estimated at 1.69 kg ha-1 (FAOSTAT, 2020).
Drought stress is the foremost limiting constraint of sub-Saharan African countries’ agriculture (Burke et al., 2010; World Bank, 2015). If one were to estimate the influence of different types of deficiency on the balance of world agricultural production, lack of water would appear to be one of the most important limiting factors. This statement is particularly true in countries located in Sub-Saharan Africa. In addition, there is still a dynamic increase in these countries’ populations (Guengant & May 2013) posing an additional challenge for local agriculture, which remains the main source of income for 90% of workers and provides more than 50% of export earnings. Thus, it becomes imperative to enhance crop productivity by increasing yield to provide sufficient food to that increasing population. Although investigations in water resources management to deal with water scarcity have been extensively conducted, drought stress remains the most important limiting factor for agricultural productivity.
Lack of fresh seed dormancy can lead to heavy crop sprouting and yield loss, particularly when terminal drought spells occur. In addition, it may be associated with heavy aflatoxin contamination as the causal fungus, Aspergillus flavus, can invade sprouting pods easily during germination compared to intact seeds (Martin et al., 1999). Therefore, as an important agronomic trait, fresh seed dormancy contributes to groundnut-based food safety I it may prevent kernels from being contaminated by aflatoxin. In general, cultivars belonging to the subspecies Arachis hypogaea subsp. hypogaea (Virginia and Runner types) have inherent dormancy, while those of the Arachis hypogaea subsp. fastigiata (Spanish and Valencia types) are extremely prone to sprouting because they usually lack fresh seed dormancy. From a breeding perspective, a few Spanish-type varieties with fresh seed dormancy have been developed from intersubspecific (i.e., Spanish x Virginia) crosses. In Sub-Saharan African countries, where most groundnut cultivars are of the Spanish type, fresh seed dormancy is a desirable trait to offset in situ germination that may occur due to late rains after groundnut maturity. However, until now, only a few Spanish varieties with fresh seed dormancy have been released. Taaru was a recent Spanish-type variety with deep fresh seed dormancy released by the ISRA groundnut breeding program in Senegal (CORAF, 2018).
In this chapter, we highlighted the main achievements in developing and releasing varieties adapted to drought-prone environments, including the potential use of high-throughput phenotyping methods and recently available groundnut genomic and genotyping resources in modern breeding approaches for improving groundnut varieties’ tolerance to drought stress. We also discussed state of the art practices in fresh seed dormancy phenotyping and genetics and pointed out the main achievements of ICRISAT and ISRA breeding programs.
An Overview of the History of Groundnut Breeding at ISRA
In Senegal, research in groundnut improvement started in 1898 when the Bambey research station was a pilot site to serve as a research platform for the French colonies in the Sudanian zone in Africa. Since then, groundnut has remained an important cash crop in Senegal. Over decades, the variety development was conditioned by the scenario of prevailing climatic conditions.
Before 1970: There was no water deficit constraint. The cv. 55-437 and 47-16 were released and widely cultivated in the northern region of Senegal (Louga). These cultivars are both 90-day maturing varieties, but the cv. 55-437 is an erect type, while cv. 47-16 is a spreading type. In the central region (Diourbel, Thiès, and Kaolack), the 120-day maturing varieties 57-422 and 48-15 were the largest cultivated varieties.
1971–1983: This period corresponded to the beginning of the drought periods in all the Sahel zone. During the 1970s, the drastic reduction of the rainfall and its erratic pattern, particularly in the Northern region of Senegal, resulted in the replacement of the cv 47-16 by the cv. 55-437 and was the only variety recommended in the Northern region (Louga). Breeding programs established during this period created a new cultivar: cv. 73-30 was released for its drought tolerance and fresh seed dormancy (Bockelee-Morvan, 1983). The late-maturing cv. 57-422 (120 days) was replaced in this large production area by the newly released cv. 73-33 that matures in 110 days. The cv. 57-422 was cornered to a small area around Diourbel.
1984–1996: This period was characterized by the release of the cv. Fleur 11, a variety with high yield potential and drought tolerance. Because of its big seed size, it has been recommended for confectionery, particularly when produced under irrigated conditions, to prevent pre-harvest aflatoxin contamination. Meanwhile, a breeding program that focuses on short duration varieties was established and resulted in the release of cv. GC-8-35 (80 days) for the replacement of cv. 55-437 in the Northern region of Senegal, where the rainy season was shortening considerably in comparison to the rest of the country. The cv. 55-422 was almost replaced by the medium duration cv. 73-33 in the Central and Southern region of the groundnut basin.
1997–2010: During this period, the breeding program on earliness also resulted in the release of four cultivars, 78-936, 73-9-11, 55-33, and SRV1-19 in 2009. Now, the cv. GC-8-35 is replaced by cv. 55-33 because the latter produces a higher haulm while maturing in 80 days. Its geographical area is expanding to the Central region of the groundnut basin.
2011–Present: In 2004, we started a new breeding program using wild species as a donor of favorable alleles. With this breeding program, chromosome segment substitution lines (CSSLs) were developed, and six were released in 2017/2018. These lines have a high yield associated with bigger seed sizes (Tossim et al., 2020), particularly those named “Tosset” and “Rafet Kaar.” These cultivars have a 100-kernel weight (60 to 65g), much higher than the recurrent parent, cv. Fleur 11. The remaining cultivars (Raw Gadu, Yaakar, Jambar, and Kom Kom) are high-yielding and show better yield stability in the Senegalese groundnut basin than Fleur 11.
Regarding fresh seed dormancy, a specific breeding program for this trait was initiated in 1997. It resulted in developing seven lines with deep fresh seed dormancy (Ndoye, 2001). However, it was only in 2018 that the line named “Taaru” was released for cultivation in Senegal for its good yield, deep fresh seed dormancy, and high oil content.
Groundnut Improvement for Drought Adaptation
1. Description of the Different Drought Scenarios and Implications in Breeding
Senegal has a decreasing rainfall gradient, ranging from less than 474 mm in the Northern groundnut basin to more than 1034 mm in the Southern region (Table 1). The upper northern groundnut basin region is drier (less than 400 mm/year), and only extra-early maturing varieties with drought tolerance traits are recommended for cultivation there. Overall, the rainfall trend shows a neat decrease since the 70s, and more frequent and severe dry spells are noted, even in Southern Senegal. Severe dry spells occurring at an early stage of the groundnut cultivation, particularly in the Northern region, might have a significant effect on productivity. There are also dry spells at later stages of the growing season, which somewhat contribute to the gap between potential crop yield and on-farm production. Therefore, both intermittent and terminal drought stress need to be considered when selecting drought tolerant varieties in Senegal.
| Agroecological Zone/Site | Average Rainfall (mm) | Growing Season Duration (days) | Occurrence of Dry Spells (lasting at least 10 days) at Early Stage |
| North Groundnut Bassin- Bambey | 487.4 | 74 | 18/40 |
| North Groundnut Bassin- Diourbel | 474.4 | 77 | 21/40 |
| South Groundnut Bassin- Kaolack | 610 | 99 | 13/40 |
| Eastern region-Sinthiou Maleme | 713 | 96 | 13/40 |
| Southern region- Kolda | 1034.3 | 105 | 14/40 |
| Agroecological Zone/Site | Duration of Drought Spells at Early Stage and Mean Standard Error | Occurrence of Dry Spells (lasting at least 10 days) at Late Stage |
Duration of Drought Spells at Late Stage and Mean Standard Error
|
| North Groundnut Bassin- Bambey | 10±1 (24) | 13/40 | 9±1 (26) |
| North Groundnut Bassin- Diourbel | 10±1 (22) | 15/40 | 9±1 (21) |
| South Groundnut Bassin- Kaolack | 9±1 (23) | 30/40 | 12±1 (20) |
| Eastern region-Sinthiou Maleme | 8±1 (16) | Dec-40 | 8±1 (14) |
| Southern region- Kolda | 8±1 (15) | 16/40 | 9±1 (17) |
Note. Values between brackets are the maximum duration of dry spells observed (Source: Service d’Agroclimatologie, ISRA/CNRA, Bambey, 2010)
To overcome the severe drought episodes in the 1970s, groundnut breeding programs in Senegal focused on developing and releasing early and drought-tolerant varieties. Three major occurrences of drought were identified and characterized: early-stage drought (after emergence or early vegetative phase), intermittent drought (at flowering and pod development stage), and terminal drought (at pod filling stage), with all types being detrimental to yield and/or its components (Clavel et al., 2007; Rao et al., 1985). Three different strategies that are related to drought tolerance have been identified in plants, including legumes (Turner et al., 2001):
Drought escapers: genotypes belonging to this class are extra-early (75–80 days) or early (90 days) maturing varieties. This drought adaptation strategy is particularly important in dry regions with frequent end-season droughts. In the northern part of Senegal, where the rainy season lasts less than 90 days, extra-early maturing varieties have been recommended for cultivation. For instance, the variety GC-8-35 was released in 1996 to replace the variety 55-437, which was previously largely cultivated in this region. Cultivar 55-437 is a 90-days maturing cultivar that was no longer getting enough rain to mature in Northern Senegal at the end of the 1990s. The breeding programs released four other extra-early maturing lines (78-936, 55-33, 73-9-11, and SRV1-19). Except for line 78-936 (an introduction), all the other ones have been developed from crosses between Chico and Senegalese elite varieties (55-437 and 73-30). The varieties 55-33 and SRV1-19 were developed from crosses between 55-437 and Chico, using a backcross and a recurrent selection scheme, respectively. These extra-early maturing varieties have higher yield performances under drought stress because they avoid drought better than CG-8-35, thanks to their physiological and membrane resistance. A breeding program is ongoing to improve these extra-early maturing lines for higher (pod and haulm) yields associated with drought tolerance traits.
Drought avoiders: genotypes that can cope with drought stress using mechanisms limiting water loss. They have different physiological mechanisms to avoid drought stress: i) a deep root system for better water uptake). This trait is critical to avoiding drought, but its limitation is a weak correlation between measurements in controlled environments and the field. With the availability of large-capacity lysimetric tubes that were recently developed at ISRA, one could investigate better the correlation of root growth parameters with physiological traits and yield under drought stress for many entries; ii) stomatal conductance (Gs) and transpiration rate (E) – in groundnut, intervarietal differences are difficult to observe even under severe drought stress. Likely, there is not much genetic variability in groundnut.
Drought tolerant: These plants would use osmotic adjustment – this mechanism has been extensively investigated in Senegal. Still, little or low genetic variability was observed between genotypes of the same phenology. Clavel et al. (2007) found that genotypic differences are greatest two weeks after drought onset.
Integrating drought adaptation traits (especially for avoidance and tolerance) in breeding programs requires high throughput phenotyping methods to screen large numbers of lines quickly.
2. Boundaries to Improve Genotypes for Drought Tolerance
Breeding for drought escape by reducing the growth duration of the varieties has been an important strategy adopted by breeding programs to tackle late water deficit stress and ensure good yields under end-season drought stress. However, breeding programs have to deal also with dry periods occurring during the plant’s developmental stage since drought spells are frequent in the Sahelian zone. The most widely used approach to evaluate genotypes’ adaptation to drought is an empirical approach that measures yield under water-limiting conditions after withholding irrigation for a given period during the developmental and reproductive stages to mimic drought spells (Clavel et al., 2007; Nigam et al., 2005). However, inconsistent climatic conditions across studies have led to contradictory statements. Besides, drought tolerance is a complex and polygenic trait.
Attempts to integrate physiological traits and yield in variety development for drought tolerance have resulted in limited success. For instance, low-cost, rapid, and easily measurable indicators for three significant physiological features of drought tolerance – the amount of water transpired (T), water-use efficiency (W), and harvest index (HI) – were developed to assess drought tolerance in crops (Devi et al., 2010; Devi et al., 2019; Tuberosa, 2012). However, application in groundnut breeding programs has been limited because of practical difficulties associated with measuring these traits under field conditions in large breeding populations. Songsri et al. (2008) found high correlations between drought tolerance index (DTI), pod yield, root length density (RLD), and harvest index (HI), indicating that RLD contributed greatly to pod yield and HI under drought conditions. This association is relevant to groundnut because pods develop underground and adequate moisture in the root zone is critical for peg and pod development. However, phenotyping for root traits in field conditions and in a large population is quite challenging.
Drought tolerance can be enhanced by improving soil water extraction and/or water use efficiency. Therefore, we recently developed a good capacity for phenotyping using a lysimetric system and high-throughput phenotyping platforms to optimize the measurements of physiological and root traits under drought stress conditions. Besides, the high throughput phenotyping methods are paving the way to solving the issue of accurate drought phenotyping in breeding programs as they usually evaluate large populations in a short period of time. Our preliminary results in this research area are quite promising as significant correlations were found between measurements of leaf temperature and leaf area index using handheld devices and with cameras embarked on drones.
3. Genetic and Genomic Resources to Breed Drought-Tolerant Genotypes
3.1 Collections
The key to success for any breeding program is the availability of a large genetic diversity for economically important traits. Characterizing and identifying the sources of variability are important steps in the pre-breeding activities of any species. In groundnut, thousands of accessions are maintained in different countries, including India (15445 accessions), the USA (9310 accessions), and China (7837 accessions) (Barkley et al., 2016). To dispose of an easily manageable and cost-effective germplasm set, it is recommended to establish a core collection (representing 10% of the overall collection) or even a mini core collection (10% of the core collection and therefore 1% of the overall collection). Thus, from the various collections kept in different countries, core collections of 1704, 831, and 576 accessions were developed and maintained respectively in India, the USA, and China (Pandey et al., 2012). From these core collections, mini core collections of 184, 112, and 298 accessions have been developed in India, the USA, and China, respectively (Jiang et al., 2010; Upadhyaya et al., 2002). Based on phenotypic data, geographic origin, and taxonomy, ICRISAT developed, as part of the Generation Challenge Program (GCP) initiative, a composite reference set of 300 genotypes (Pandey et al., 2012). Recently, in the framework of the Peanut Innovation Lab (PIL), a USAID Feed the Future initiative, ISRA has assembled a collection of 1049 accessions from nine countries in West, East, and Southern Africa, in partnership with breeders from national research programs of these countries. These accessions were genotyped at the University of Georgia (UGA) using the Affymetrix chip (45K SNP markers). The genotypic data have been made publicly available at https://www.peanutbase.org. From that germplasm, a subset of 300 accessions representing the whole genetic diversity in the entire collection was established using both the genotypic data and breeders’ knowledge. That subset is characterized in all the nine germplasm-contributing countries for various traits, including yield, drought tolerance, and fresh seed dormancy. The phenotypic data generated will be used for association mapping studies to better understand the genetics and mechanisms involved in drought tolerance in groundnut.
Because of the narrow genetic base of groundnut, compared to other crop species, wild species related to cultivated groundnut represent valuable sources of useful alleles essential for groundnut improvement. Major centers holding wild Arachis species collections include Texas A&M University (1200 accessions), U.S. Department of Agriculture (USDA, 607 accessions), North Carolina State University, (NCSU, 406 accessions), Brazil EMBRAPA-CENARGEN (1220 accessions), ICRISAT in India (477 accessions) and the Instituto de Botánica del Nordeste (IBONE, 472 accessions) in Argentina (Pandey et al., 2012). In addition, breeding programs worldwide have developed different types of biparental populations, including recombinant inbred lines, advanced backcross QTL lines, and chromosome segment substitution lines (Foncéka et al., 2012; Table 2). These genetic materials could be useful sources for identifying QTLs involved in drought tolerance using high-throughput genotyping and accurate and rapid phenotyping approaches.
| Population | Lines/Types | Segregating Traits |
Research Institute /University
|
| A. duranensis × A. stenosperma | 87 RILs | Late leaf spot resistance, transpiration, response to drought stress, various aspects of plant morphology | EMBRAPA |
| A. hypogaea cv. Fleur11 × (A. ipäensis × A. duranensis) | 81 BC4F7 | Seed size and pod and haulm yield, disease resistance |
ISRA (CERAAS), CIRAD
|
| A. hypogaea cv. Fleur11 × (A. batizocoi×A. duranensis) | 133 BC2F7 | Seed size and pod and haulm yield, disease resistance |
ISRA (CERAAS)
|
| A. hypogaea cv. Fleur11 × (A. valida×A. duranensis) | 300 BC2F7 | Seed size and pod and haulm yield, disease resistance |
ISRA (CERAAS)
|
| Tifrunner × C76-16, Florida-07 × C76-16 | 400 RILs | Drought tolerance and reduced PAC |
USDA-ARS, UGA, NCSU
|
| TAG 24 × ICGV 86031 | 318 RILs | Drought related traits | ICRISAT |
| ICGS 44 × ICGS 76 | 188 RILs | Drought related traits | ICRISAT |
| ICGS 76 × CSMG 84-1 | 176 RILs | Drought related traits | ICRISAT |
3.2 Genomics Resources
Genomic resources are powerful tools to harness the genetic variation in germplasm collections for the faster generation of improved varieties. The groundnut scientific community has made a considerable leap from insufficient to abundant genomic resources in this area. Multiple publications provide details on the available resources (Ozias-Akins et al., 2017; Pandey et al., 2012; Pandey et al., 2014; Varshney et al., 2014; Vishwakarma et al., 2017). All these tools and technologies are useful for basic and applied groundnut improvement research. In addition to the various molecular markers and genetic maps, the genome sequences of the wild diploid parents of cultivated groundnut (Arachis ipaensis and Arachis duranensis) and the tetraploid variety Tifrunner (Bertioli et al., 2016; Chen et al., 2016) are valuable resources. Sequencing the reference genome is one of the most important milestones for any crop species to accelerate the process of understanding genome architecture, gene discovery and molecular breeding for complex traits such as drought. Available groundnut genome sequences paved the way for the development and deployment of highly informative Affymetrix technology Single Nucleotide Polymorphism (SNP) chips “Axiom_Arachis” containing 58k and 48k markers (Clevenger et al., 2017; Pandey et al., 2017). Genome sequencing has also enabled the development of more simple sequence repeats (SSR) and insertions/deletion (INDELs) markers (Vishwakarma et al., 2017; Zhao et al., 2017). After completing the reference genome sequences, a great challenge for researchers is understanding the functions of the entire sets of genes. The gene expression atlas of groundnut provides detailed information on gene expression in different genotypes, organs, tissues, cells, and developmental stages (Pandey & Varshney, 2018). Such resources were developed by the University of Georgia using RNA sequencing data generated from 22 tissues (Clevenger et al., 2017). ICRISAT also reported the development of another gene expression atlas (AhGEA) using RNA-Seq data of 19 tissues from five stages of an early maturing, high-yielding, drought-tolerant groundnut variety, ICGV 91114 (Pandey & Varshney, 2018). These resources could be useful for conducting high-resolution trait mapping, marker development, candidate gene discovery, and molecular breeding for drought tolerance in groundnut.
Moreover, several QTLs are identified for drought tolerance traits, but their validation and application in marker-assisted selection (MAS) are challenging (Foncéka et al., 2012; Faye et al., 2015; Gautami et al., 2012; Ravi et al., 2011). These authors suggested that marker-assisted recurrent selection or genomic selection (GS) would be more appropriate in order to combine favorable alleles from so many regions. GS-assisted breeding has been tested in groundnut using a training population (TP) of 340 advanced breeding lines followed by multi-seasons and multi-locations phenotyping for important agronomic traits (Janila et al., 2016a). Currently, few successful examples of molecular breeding products are available in groundnut (Chu et al., 2011; Janila et al., 2016b; Varshney et al., 2014). With the greater development of genetic resources and advanced genomic tools, the potential to develop drought-tolerant cultivars is greater than ever. New emerging breeding technologies and methods such as genomic selection and genome editing are promising tools. Genetic engineering and genome editing have great potential in improving groundnut for drought tolerance as such technologies are not limited by ploidy and cross ability barriers. Several research groups, particularly in the United States, China, and India, have developed transgenic groundnut lines to resist viruses, insects, fungi, as well as for drought tolerance, and grain quality. Brasileiro et al. (2014) reviewed the progress achieved in Arachis genetic transformation. Tolerance was reported in drought-susceptible groundnut variety JL24 by over-expressing the gene DREB1A (Bhatnagar-Mathur et al., 2008). This transgenic event improved groundnut yield through a complex molecular mechanism involving a cascade of stress signaling and growth-regulating genes. However, in most West African countries, particularly Senegal, limited by the availability of research funds, suitable infrastructures, and regulatory approval, the use of genetic engineering tools remains uncertain. In such conditions, crop modeling is more applicable, especially for traits that are difficult to phenotype, such as physiological traits.
Groundnut Improvement for Seed Dormancy
In the Sahelian zone, late rains after groundnut’s physiological maturity can cause important yield losses (20 and 50%) in varieties lacking fresh seed dormancy (Gautreau, 1984; Khalfaoui, 1991; Ndoye, 2001). In addition, the sprouting of non-dormant varieties can increase aflatoxin contamination of kernels (Martin et al., 1999). Thus, there is a need to develop Spanish–type varieties with fresh seed dormancy to improve groundnut productivity and seed quality for areas prone to late rains.
1. Development and Release of Dormant Varieties
ICGV 86155, ICGV 86156, ICGV 86158, ICGV 87378, and ICGV 87921 Spanish groundnut (Arachis hypogaea L. subsp. fastigiata Waldron var. vulgaris Harz) improved germplasm lines (Reg. no. GP-79 to GP-83, P1 594969 to P1 594973), bred by ICRISAT, India, were released in 1995 for their fresh seed dormancy in the Spanish background (Upadhyaya et al., 1997). Another improved Spanish peanut (Arachis hypogaea L. subsp. fastigiata var. vulgaris) germplasm bred by ICRISAT, ICGV 93470 (Reg. no. GP-102, PI 614087) was released in 1999 for its early-maturity and fresh seed dormancy (Upadhyaya et al., 2001).
In Senegal, the first Spanish dormant variety released is the variety 73-30 (Bockelee-Morvan, 1983). It has deep dormancy and could be used as a donor parent in breeding programs for early maturing and dormant groundnut varieties. A second dormant Spanish variety derived from a cross between the variety Fleur 11 and 73-30 was recently released. That new variety named “Taaru” has a bigger seed size than variety 73-30, while its oil content is higher (53%) than most of the Senegalese groundnut varieties.
Screening of various groundnut germplasm, including core collections, should allow the identification of new sources of fresh seed dormancy for further varietal improvement. In our breeding program, we recently evaluated the above-mentioned collection of 289 accessions for fresh seed dormancy. Among the 189 short-duration cycle accessions, 16 were classified as having deep dormancy (percentage of germinated seeds < 10%), 23 were classified as having light dormancy (< 30%), and the remaining 150 accessions were classified as non-dormant (percentage of germinated seeds > 30%). Out of the 100 long-duration cycle accessions, 40 accessions were classified with deep dormancy, 26 accessions with light dormancy, and 34 accessions as non-dormant (Unpublished data). Some accessions with fresh seed dormancy are listed below (Table 3). This germplasm is an important source of accessions with fresh seed dormancy.
| Duration Cycle Length | Origin /Country /Maintainer | Accessions |
Other Characteristics
|
| Short | Senegal/ISRA | 73-30 |
Drought tolerance
|
| Short | Senegal/ISRA | Taaru |
High oleic content (53%)
|
| Long | Senegal/ISRA | 69-101 |
Leaf spot & Rosette resistance
|
| Long | Senegal/ISRA | Turquie 2 |
Confectionery type and mid-oleic content
|
| Short | Ghana/CSIR-SARI | ICGV 13110 | ND |
| Short | Ghana/CSIR-SARI | ICGV 15017 | ND |
| Long | Ghana/CSIR-SARI | NUMEX 05 | High yield |
| Short | Uganda/NARO | SVG-GB-10010 | ND |
| Short | Uganda/NARO | ICGV SM 08577 | ND |
| Short | Malawi/DAR | ICGV SM 5738 | ND |
| Short | Malawi/DAR | ICGV 6057 | ND |
| Short | Malawi/DAR | ICGV SM07544 | ND |
| Short | Mozambique/IIAM | Pan 13005 | ND |
| Short | Mozambique/IIAM | ICGV-SM 03530 | ND |
Note. ND: not determined
2. Phenotyping for Fresh Seed Dormancy
Assigning a phenotypic value to every individual of the population is critical to achieving a genetic gain in any breeding program; in other words, assuming the same selection intensity and population variance, the higher the heritability, the more important the genetic gain. However, because heritability is somewhat a matter of accuracy of the phenotyping approach, biases can sometimes be misleading. Phenotyping for seed dormancy can be done either by evaluating the in situ number of sprouts or weight and expressing it as a percentage of the total number or mass of harvested seeds or by performing an in vitro seed germination test (Faye et al., 2010; Gautreau, 1984; Khalfaoui, 1991; Ndoye, 2001; Upadhyaya & Nigam, 1999) using fresh or cured seeds. Considering the above statements related to dormancy phenotyping, it seems necessary to highlight tips that can improve phenotyping and, subsequently, the selection of dormant cultivars. Firstly, one needs to make sure the seeds are fully mature. Due to its indeterminate nature of flowering, seed maturity level may vary from one pod to another for the same individual plant. A simple method for selecting mature seeds is the observation of the black coloration of the hull’s internal parenchyma (Miller & Burns, 1971). This parenchyma turns brown to black when the pod is fully mature and white to yellow if the pod is immature. Secondly, as the seed coat and cotyledons and embryos have a role in imparting dormancy in groundnut (Nautiyal et al., 1994), care should be taken when shelling pods for the in vitro seed germination tests. Thirdly, two-seeded pods have deeper dormancy at their basal seed than at their apical seed (Ketring & Morgan, 1971). Therefore, it seems more relevant to use only apical seeds when phenotyping for fresh seed dormancy to harmonize sampling.
3. Genetic Improvement for Seed Dormancy
Various genetic studies were carried out to understand the inheritance of fresh seed dormancy in peanuts (Asibuo et al., 2008; Faye et al., 2010; Gautreau, 1984; Khalfaoui, 1991; Lin & Lin, 1971; Ndoye, 2001; Upadhyaya & Nigam, 1999). Some studies revealed that fresh seed dormancy is controlled by a dominant allele of a single gene (Asibuo et al., 2008; Faye et al., 2010; Upadhyaya & Nigam, 1999). In contrary, other studies (Khalfaoui, 1991; Ndoye, 2001) reported polygenic inheritance with epistasic effects. More recently, Vishwakarma et al. (2016) found that the trait is controlled by two recessive genes. These converses may be due to the different phenotyping methods and the nature of the crosses used by authors. Some crosses were intra-subspecific (Spanish × Spanish), while others were inter-subspecific (Spanish × Virginia). In most of the studies of seed dormancy genetics in groundnut, authors concluded that broad sense heritability was moderate to high (<0.50), indicating the involvement of both significant genetic and environmental factors (Faye et al., 2010; Khalfaoui, 1991; Ndoye, 2001). In addition, significant genotype × year effects were reported (Ndoye, 2001).
4. Toward the Identification of Molecular Markers Linked to Seed Dormancy in Groundnut
Vishwakarma et al. (2016) identified two major QTLs (qfsd-1 and qfsd-2) for fresh seed dormancy in the linkage groups A05 and B06, respectively. Using a recombinant inbred line (RIL) mapping population derived from a cross between Tifrunner (a dormant Runner type) and GT-C20 (a non-dormant Spanish type), Wang et al. (2022) identified two major seed dormancy QTLs on chromosome A04 and A05 with 43.2% and 51.6% of the phenotype variation explained, respectively. Besides, a candidate gene (Arahy.KB746A) coding and ethylene-responsive transcription factor was found very close to the QTL mapped on chromosome A05. Deploying the QTL-seq approach, Kumar et al. (2020) identified other key genomic regions and candidate genes for fresh seed dormancy on the B05 and A09 pseudomolecules. They developed a validated marker (GMFSD1) that can be used in MAS programs to improve seed dormancy in groundnut.
In our breeding program in Senegal, we developed two RILs populations for fresh seed dormancy. One of the RILs populations is derived from a cross between cv. Fleur 11 and cv. 73-30, while the other is derived from a cross between ICGV 96894 and cv. 73-30. We identified eight lines with deep dormancy, 3 RILs (C1P15-18-4, C1P24-1-1, C1P18-6-5, and C1P30-9-8) of the population Fleur 11 × 73-30, and 4 RILs (C3P11-8-2; C3PNI-1-3, C3P7-1-1, and C3P6-2-5) from the cross ICGV 96894 × 73-30 (Figure 1). Further analyses are ongoing for the mapping of seed dormancy QTLs in these two populations.
Figure 1
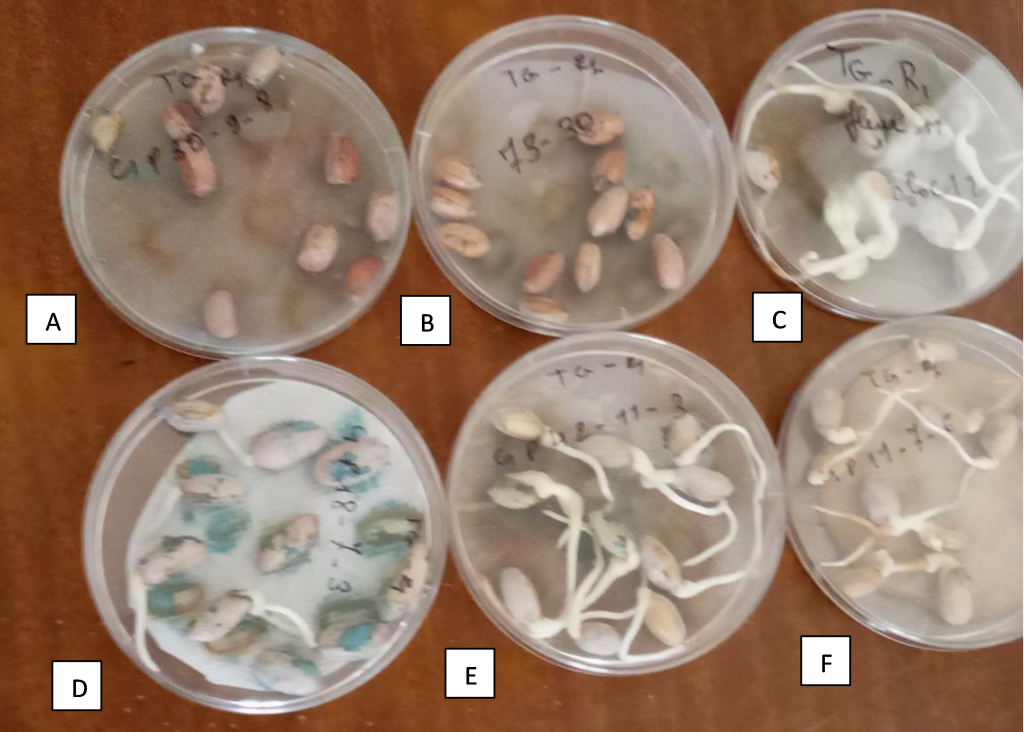
Note. (A): C1P30-9-8, a line with deep fresh seed dormancy; (B): cv 73-30, the dormant parental line; (C): cv Fleur 11, the non-dormant parental line; (D), (E) and (F) are non-dormant recombinant inbred lines.
Conclusion
In Senegal, the groundnut breeding program was established in the early 20th century. Among its great achievements is the development and registration of very early-maturing varieties (75–80 days), among which cv. 55-33 and GC-8-35 are widely cultivated in the Northern region of Senegal where the rainfall is particularly low and the rainy season is very short (less than three months). Similarly, a clear shortening of the rainy season in the southern region of the Groundnut Basin has led to the release of Spanish varieties maturing 90-days after planting, among which is «Taaru», a variety with 2-3 weeks post maturity fresh seed dormancy. Other short-duration varieties with bigger grain sizes than previous varieties were developed (Raw Gadu, Rafet Kaar, Yaakar, Tosset, Jaambar, and Kom Kom) using wild introgressions from the synthetic (Arachis duranensis × Arachis ipaënsis). As wild-related species have been proven effective in improving groundnut productivity under harsh conditions such as drought and diseases, new crosses, including other wild-related species, have been initiated to develop advanced backcross populations. Beyond the long-term objective of releasing new varieties, those populations could be useful in deciphering physiological and genetic traits related to drought tolerance in groundnut. On another hand, breeding for short-duration varieties with fresh seed dormancy is necessary to facilitate farmers’ wide adoption of these varieties. Therefore, it is critical to consider fresh seed dormancy as a key selection trait, particularly when breeding groundnut for Sahelian and Sudano-Sahelian zones. With the fast development of high throughput phenotyping platforms and the availability of groundnut genomic resources, significant progress could be achieved toward higher genetic gains for all the traits contributing to yield and seed quality in groundnut under marginal environments.
References
ANSD, Agence Nationale de la Statistique et de la Démographie (2022). Situation économique et sociale du Sénégal 2019. Rapport d’activité de l’ANSD, 310 pages.
Asibuo, A. Y., Richard, A., Osei, S. -K., Dapaah, K. H., Seth, O. -D., & Adelaide, A. (2008). Inheritance of fresh seed dormancy in groundnut. African Journal of Biotechnology, 7(4), 421–424.
Barkley, N. A., Upadhyaya, H. D., Liao, B., & Holbrook, C. C. (2016). Global resources of genetic diversity in peanut. In Stalker, H. T. & Wilson R. F. (Eds), Peanuts, AOCS Press (pp 67–109). https://doi.org/10.1016/B978-1-63067-038-2.00003-4.
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., Liu, X., Gao, D., Clevenger, J., Dash, S., Ren, L., Moretzsohn, M. C., Shirasawa, K., Huang, W., Vidigal, B., Abernathy, B., & Chu, Y. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nature Genetics, 48, 438–446. https://doi.org/10.1038/ng.3517
Bhatnagar-Mathur, P., Vadez, V., & Sharma, K. K. (2008). Transgenic approaches for abiotic stress tolerance in plants: Retrospect and prospects. Plant Cell Reports, 27(3), 411–424. https://doi.org/10.1007/s00299-007-0474-9
Brasileiro, M., Araujo, G., Leal-Bertioli, A. C., & Guimarães, S. C. (2014). Genomics and genetic transformation in Arachis. International Journal of Plant Biology, 2(3), 1017.
Bockelee-Morvan, A. (1983). Les différentes variétés d’arachide : Répartition géographique et climatique, disponibilité. Oléagineux, 38(2), 73–116.
Burke, M., de Janvry, A., & Quintero, J. (2010). Providing index-based agricultural insurance to smallholders: Recent progress and future promise. CEGA University of California at Berkeley: Berkeley, CA, USA. https://web.worldbank.org/archive/website01589/WEB/IMAGES/PARAL-18.PDF
Chen, X., Li, H., Pandey, M. K., Yang, Q., Wang, X., Garg, V., & Li, H. (2016). Draft genome of the A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proceedings of the National Academy of Sciences of the Uni- Ted States of America 113, 6785–6790. https://doi.org/10.1073/pnas.1600899113
Chu, Y., Wu, C. L., Holbrook, C. C., Tillman, B. L., & Person, G. (2011). Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome, 4, 110–117. https://doi.org/10.3835/plantgenome2011.01.0001
Clavel, D., Baradat, P., Khalfaoui, J. L., Drame, N. K., Diop, N. D., Diouf, O., Zuily Fodil, Y. (2007). Adaptation à la sécheresse et création variétale : Le cas de l’arachide en zone sahélienne Deuxième partie : Une approche pluridisciplinaire pour la création variétale. OCL, 14 (5), 293–308. https://doi.org/10.1051/ocl.2007.0141
Clevenger, J., Chu, Y., Chavarro, C., Agarwal, G., Bertioli, D. J., Leal-Bertioli, S. C. M., Pandey, M. K., Vaughn, J., Abernathy, B., Barkley, N. A., Hovav, R., Burow, M., Nayak, S. N., Chitikineni, A., Isleib, T. G., Holbrook, C. C., Jackson, S. A., Varshney, R. K., & Ozias-Akins, P. (2017). Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Molecular Plant, 10(2), 309–322. https://doi.org/10.1016/j.molp.2016.11.015
CORAF (2018). Catalogue régional des espèces et variétés végétales CEDEAO-UEMOA-CILSS: Variétés homologuées 2016-2018, 27 pages.
Devi, J. M., Sinclair, T. R., & Vadez, V. (2010). Genotypic variation in peanut for transpiration response to vapor pressure deficit. Crop Science, 50(1), 191–196. https://doi.org/10.2135/cropsci2009.04.0220
Devi, J. M., Sinclair, T. R., Vadez, V., Shekoofa, A., & Puppala, N. (2019). Strategies to Enhance Drought Tolerance in Peanut and Molecular Markers for Crop Improvement, Vol. II. Springer International Publishing. https://doi.org/10.1007/978-3-319-99573-1_8
FAOSTAT (2020). Production-cultures et produits animaux. www.fao.org/faostat/fr/#compare.
Faye, I., Foncéka, D., Rami, J. F., Tossim, H. A, Sall, M. N., Diop, A. T. & Ndoye, O. (2010). Inheritance of fresh seed dormancy in Spanish-type peanut (Arachis hypogaea L.): bias introduced by inadvertent selfed flowers as revealed by microsatellite markers control. African Journal of Biotechnology, 9(13), 1905–1910.
Faye, I., Pandey, M. K., Hamidou, F., Rathore, A., Ndoye, O., Vadez, V., & Varshney, R. K. (2015). Identification of quantitative trait loci for yield and yield related traits in groundnut (Arachis hypogaea L.) under different water regimes in Niger and Senegal. Euphytica, 206(3), 631–647. https://doi.org/10.1007/s10681-015-1472-6
Foncéka, D., Tossim, H. A., Rivallan, R., Vignes, H., Lacut, E., de Bellis, F., Faye, I., Ndoye, O., Leal-Bertioli, S. C. M., Valls, J. F. M., Bertioli, D. J., Glaszmann, J. C., Courtois, B., & Rami, J. F. (2012). Construction of chromosome segment substitution lines in peanut (Arachis hypogaea L.) using a wild synthetic and QTL mapping for plant morphology. PLoS ONE, 7(11). https://doi.org/10.1371/journal.pone.0048642
Gautami, B., Pandey, M. K., Vadez, V., Nigam, S. N., Ratnakumar, P., Krishnamurthy, L., Radhakrishnan, T., Gowda, M. V. C., Narasu, M. L., Hoisington, D. A, Knapp, S. J., & Varshney, R. K. (2012). Quantitative trait locus analysis and construction of consensus genetic map for drought tolerance traits based on three recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.). Molecular Breeding, 30, 757–772. https://doi.org/10.1007/s11032-011-9660-0
Gautreau J., (1984). Evaluation des taux effectifs de non-dormance au champ d’arachides sénégalaises. Oléagineux, 39(2), 83–88.
Guengant, J. P., & May, J. F. (2013). Africa 2050: Background paper on African demography. https://www.emergingmarketsforum.org/wp-content/uploads/2020/01/Final-Program-min-1.pdf
Janila, P, Variath, M. T., Pandey, M. K., Desmae, H., & Motagi, B. N. (2016a). Genomic tools in groundnut breeding program: Status and perspectives. Frontiers in Plant Science, 7(March), 2004–2013. https://doi.org/10.3389/fpls.2016.00289
Janila, P., Pandey, M. K., Shasidhar, Y., Variath, M. T., Sriswathi, M., Khera, P., Manohar, S. S., Nagesh, P., Vishwakarma, M. K., Mishra, G. P., Radhakrishnan, T., Manivannan, N., Dobariya, K. L., Vasanthi, R. P., & Varshney, R. K. (2016b). Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing genotypes. Plant Science, 242, 203–213. https://doi.org/10.1016/j.plantsci.2015.08.013
Jiang, H. F., Ren, X. P., Zhang, X. J., Huang, J. Q., Lei, Y., Yan, L. Y., Liao, B. S., Upadhyaya, H. D., & Holbrook, C. C. (2010). Comparison of genetic diversity based on SSR markers between mini core collections from China and ICRISAT. Acta Agronomica Sinica, 36(7), 1084–1091. https://doi.org/10.1016/S1875-2780(09)60059-6
Ketring, D. L., & Morgan, P. W. (1971). Physiology of oilseeds. II. Dormancy release in Virginia-type peanut seeds by plant growth regulators. Plant Physiology, 47(4), 488–492. https://doi.org/10.1104/pp.47.4.488
Khalfaoui, J. -L. B. (1991). Inheritance of seed dormancy in a cross between two Spanish peanut cultivars. Peanut Science, 18(2), 65–67. https://doi.org/10.3146/i0095-3679-18-2-1
Kumar, R., Janila, P., Vishwakarma, M. K., Khan, A.W., Manohar, S. S., Gangurde, S. S., Variath, M. T., Shasidhar, Y., Pandey, M. K., & Varshney, R. K. (2020). Whole-genome resequencing-based QTL-seq identified candidate genes and molecular markers for fresh seed dormancy in groundnut. Plant Biotechnology Journal 18(4): 992–1003. https://doi.org/10.1111/pbi.13266.
Lin, H., & Lin, C. Y., (1971). Studies on the seed dormancy of peanuts. III. Inheritance of seed dormancy of peanuts. Journal of Agricultural Research, 20, 49–53.
Martin J., Ba, A., Dimanche, P., & Schilling, R. (1999). Comment lutter contre la contamination de l’arachide par les aflatoxines ? Expériences conduites au Sénégal. Agriculture et Développement, 23, 58–67.
Miller, O. H. & Burns, E. E. (1971). Internal color of Spanish peanut hulls as an index of kernel maturity. Journal of Food Science, 36(4), 669–670. https://doi.org/10.1111/j.1365-2621.1971.tb15157.x
Nautiyal, P. C., Bandyopadhyay, A., & Zala, P. V. (2001). In situ sprouting and regulation of fresh-seed dormancy in Spanish-type groundnut (Arachis hypoagaea L.). Field Crops Research, 70(3), 233–241. https://doi.org/10.1016/S0378-4290(01)00143-5
Ndoye, O. (2001). Screening techniques and mode of inheritance of fresh seed dormancy among crosses of Spanish-type peanut (Arachis hypogaea L.). [Doctoral dissertation, Texas A&M University], Texas A&M University ProQuest Dissertations Publishing, 2001. 3020100, 162 pages.
Nigam, S. N., Chandra, S., Rupa Sridevi, K., Bhukta, M., Reddy, A. G. S., Rachaputi, N. R., Wright, G. C., Reddy, P. V., Deshmukh, M. P., Mathur, R. K., Basu, M. S., Vasundhara, S., Vindhiya Varman, P., & Nagda, A. K. (2005). Efficiency of physiological trait-based and empirical selection approaches for drought tolerance in groundnut. Annals of Applied Biology, 146(4), 433–439. https://doi.org/10.1111/j.1744-7348.2005.040076.x
Ozias-Akins, P., Cannon, E. K. S., & Cannon, S. B. (2017). Genomics resources for improvement. In Varshney, R., Pandey, M., Puppala, N. (Eds.), The peanut genome. (pp. 69–91). https://doi.org/10.1007/978-3-319-63935-2_6
Pandey, M. K., Agarwal, G., Kale, S. M., Clevenger, J., Nayak, S. N., Sriswathi, M., Chitikineni, A., Chavarro, C., Chen, X., Upadhyaya, H. D., Vishwakarma, M. K., Leal-Bertioli, S., Liang, X., Bertioli, D. J., Guo, B., Jackson, S. A., Ozias-Akins, P., & Varshney, R. K. (2017). Development and evaluation of a high density genotyping “Axiom-Arachis” array with 58 K SNPs for accelerating genetics and breeding in groundnut. Scientific Reports, 7(December 2016), 1–10. https://doi.org/10.1038/srep40577
Pandey, M. K., Monyo, E., Ozias-Akins, P., Liang, X., Guimarães, P., Nigam, S. N., Upadhyaya, H. D., Janila, P., Zhang, X., Guo, B., Cook, D. R., Bertioli, D. J., Michelmore, R., & Varshney, R. K. (2012). Advances in Arachis genomics for improvement. Biotechnology Advances, 30(3), 639–651. https://doi.org/10.1016/j.biotechadv.2011.11.001
Pandey, M. K., Upadhyaya, H. D., Rathore, A., Vadez, V., Sheshshayee, M. S., Sriswathi, M., Govil, M., Kumar, A., Gowda, M. V. C., Sharma, S., Hamidou, F., Kumar, V. A., Khera, P., Bhat, R. S., Khan, A. W., Singh, S., Li, H., Monyo, E., Nadaf, H. L., & Varshney, R. K. (2014). Genome wide association studies for 50 agronomic traits in using the “reference set” comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PloS One, 9(8), e105228. https://doi.org/10.1371/journal.pone.0105228
Pandey, M. K., & Varshney, K. R. (2018). Groundnut entered post-genome sequencing era: Opportunities and challenges in translating genomic information from genome to field. In S. H. W. S. S. Gosal (Ed.), Biotechnologies of Crop Improvement (pp. 199–109). Springer International Publishing. https://doi.org/https://doi.org/10.1007/978-3-319-94746-4_9
Rao, R. C. N., Singh, S., Sivakumar, M. V. K., Srivastava, K. L., & Williams, J. H. (1985). Effect of water deficit at different growth phases of peanut. I. yield responses. Agronomy Journal, 77(5), 782–786. https://doi.org/10.2134/agronj1985.00021962007700050026x
Ravi, K., Vadez, V., Isobe, S., Mir, R. R., Guo, Y., Nigam, S. N., Gowda, M. V. C., Radhakrishnan, T., Bertioli, D. J., Knapp, S. J., & Varshney, R. K. (2011). Identification of several small main-effect QTLs and a large number of epistatic QTLs for drought tolerance related traits in groundnut (Arachis hypogaea L.). Theoretical and Applied Genetics, 122, 1119–1132. https://doi.org/10.1007/s00122-010-1517-0
Songsri, P., Jogloy, S., Vorasoot, N., Akkasaeng, C., Patanothai, A., & Holbrook, C. C. (2008). Root distribution of drought-resistant genotypes in response to drought. Journal of Agronomy and Crop Science, 194(2), 92–103. https://doi.org/10.1111/j.1439-037X.2008.00296.x
Tossim, H. A., Nguepjop, J. R., Diatta, C., Sambou, A., Seye, M., Sane, D., Rami, J. F., & Foncéka, D. (2020). Assessment of 16 peanut (Arachis hypogaea L.) CSSLs derived from an interspecific cross for yield and yield component traits: QTL validation. Agronomy, 10(4), 583. https://doi.org/10.3390/agronomy10040583
Tuberosa, R. (2012). Phenotyping for drought tolerance of crops in the genomics era. Frontiers in Physiology, 3(September 2012), 1–26. https://doi.org/10.3389/fphys.2012.00347.
Turner, N. C, Wright, G. C., Siddique, K. (2001). Adaptation of grain legumes to water-limited environments. Advances in Agronomy, 71, 193–231. https://doi.org/10.1016/S0065-2113(01)71015-2
Upadhyaya H. D., & Nigam N. S. (1999). Inheritance of fresh seed dormancy in peanut. Crop Science, 39(1), 98–101. https://doi.org/10.2135/cropsci1999.0011183X003900010015x
Upadhyaya, H. D., Nigam, S. N., Rao, J. V., Reddy, A. G. S., Yellaiah, Y. & Reddy, N. S. (1997). Registration of five Spanish peanut germplasm lines with fresh seed dormancy. Crop Science, 37(3), 1027–1027. https://doi.org/10.2135/cropsci1997.0011183X003700030088x
Upadhyaya, H. D., Nigam, S. N., Reddy, A. G. S. & Yellaiah, N. (2001). Registration of early-maturing fresh seed dormant peanut germplasm ICGV 93470. Crop Science, 41(2), 597–598. https://doi.org/10.2135/cropsci2001.412597-ax
Upadhyaya, H. D., Bramel, P. J., Ortiz, R., & Singh, S. (2002). Developing a mini core of peanut for utilization of genetic resources. Crop Science, 42(6), 2150–2156. https://doi.org/10.2135/cropsci2002.2150
Varshney, R. K., Pandey, M. K., Janila, P., Nigam, S. N., Sudini, H., Gowda, M. V. C., Sriswathi, M., Radhakrishnan, T., Manohar, S. S., & Nagesh, P. (2014). Marker assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theoretical and Applied Genetics, 127(8). https://doi.org/10.1007/s00122-014-2338-3.
Vishwakarma, M. K., Pandey, M. K., Shasidhar, Y., Manohar, S. S., Nagesh, P., Janila, P., & Varshney, R. K. (2016). Identification of two major quantitative trait locus for fresh seed dormancy using the diversity arrays technology and diversity arrays technology-seq based genetic map in Spanish-type peanuts. Plant Breeding, 135(3), 367–375. https://doi.org/10.1111/pbr.12360
Vishwakarma, M. K., Kale, S. M., Sriswathi, M., Naresh, T., Shasidhar, Y., Garg, V., Pandey, M. K., & Varshney, R. K. (2017). Genome-wide discovery and deployment of insertions and deletions markers provided greater insights on species, genomes, and sections relationships in the genus arachis. Frontiers in Plant Science, 8(December). https://doi.org/10.3389/fpls.2017.02064
Wang, M. L., Wang, H., Zhao, C., Tonnis, B., Tallury, S., Wang, X., Clevenger J. & Guo B. (2022). Identification of QTLs for seed dormancy in cultivated peanut using a recombinant inbred line mapping population. Plant Molecular Biology Reports 40, 208–217. https://doi.org/10.1007/s11105-021-01315-5
World Bank. (2015). Groundnut value chain competitiveness and prospects for development. The World Bank Agriculture Global Practice – West Africa (GFA01) Africa Region, 50 pages.
Zhao, C., Qiu, J., Agarwal, G., Wang, J., Ren, X., Xia, H., Guo, B., Ma, C., Wan, S., Bertioli, D. J., Varshney, R. K., Pandey, M. K., & Wang, X. (2017). Genome-wide discovery of microsatellite markers from diploid progenitor species, arachis duranensis and A. Ipaensis, and their application in cultivated peanut (A. hypogaea). Frontiers in Plant Science, 8(July), 1–12. https://doi.org/10.3389/fpls.2017.01209

